16 Oct Dactylonectria
Dactylonectria L. Lombard & Crous, in Lombard et al., Phytopath. Mediterr. 53(3): 523 (2014)
The genus Dactylonectria was introduced by Lombard et al. (2014) for a group of species which were previously treated in Ilyonectria (Chaverri et al. 2011; Cabral et al. 2012a, b, c). In morphology, Dactylonectria resembles Ilyonectria and Neonectria but can be distinguished by their characteristic ovoid to obpyriform, smooth to finely warted, dark-red ascomata with a papillate ostiolar region at the apex (Lombard et al. 2014; Gordillo and Decock 2018). Species of this genus are mostly associated with Vitis sp., while some species are also recorded from other hosts (Farr and Rossman 2019).
Classification – Sordariomycetes, Hypocreomycetidae, Hypocreales, Nectriaceae
Type species – Dactylonectria macrodidyma (Hallen, Schroers & Crous) L. Lombard & Crous, in Lombard et al., Phytopath. Mediterr. 53(3): 527 (2014)
Distribution – Worldwide
Disease symptoms – Black foot disease, black root rot
Characteristic symptoms of black foot disease include a reduction in root biomass and root hairs with sunken and necrotic lesions (Hallen et al. 2006). Severe necrosis of the root system results in stunting, wilting, leaf chlorosis, browning and leaf drop prior to death (Parkinson et al. 2017). Dactylonectria alcacerensis, D. estremocensis, D. macrodidyma, D. novozelandica, D. pauciseptata, D. pinicola, D. torresensis, and D. vitis are associated with black foot disease of grapevine (Cabral et al. 2012a; Lombard et al. 2014).
Hosts – Abies sp., Annona cherimola, Anthrium sp., Arbutus unedo, Cistus albidus, Crataegus azalous, Erica melanthera, Eriobotrya japonica, Ficus sp., Fragaria sp., Hordeum vulgare, Ilex aquifolium, Juglans regia, Juniperus phoenicea, Lonicera sp., Myrtus communis, Persea americana, Picea glauca, Pinus sp., Pistacia lentiscus, Prunus domestica, Pyracantha sp., Quercus sp., Rosmarinus officinalis, Santolina chamaecyparissus and Vitis sp.
Fig. Symptoms of black foot disease on Vitis spp. a, b. dead plants and stunted growth of grapevine. c, e. infection of rootstock d. blocked xylem vessels f. black streak (Courtesy of Halleen).
Morphological based identification and diversity
Lombard et al. (2014) accepted ten species in Dactylonectria based on ITS, LSU, TUB2 and tef1 sequence data and morphological characters. Later, Gordillo and Decock (2018) introduced another four species to the genus based on morphology and sequence data.
Fourteen Dactylonectria species have been described with DNA sequence data in GenBank. Dactylonectria species produce cylindrocarpon-like asexual morphs, several of which were previously treated in Ilyonectria (Lombard et al. 2014). Dactylonectria species were distinguished mainly by phylogenetic inference and using unique fixed single nucleotide polymorphisms (SNP’s) rather than morphological characters (Lombard et al. 2014; Gordillo and Decock 2018).
Molecular based identification and diversity
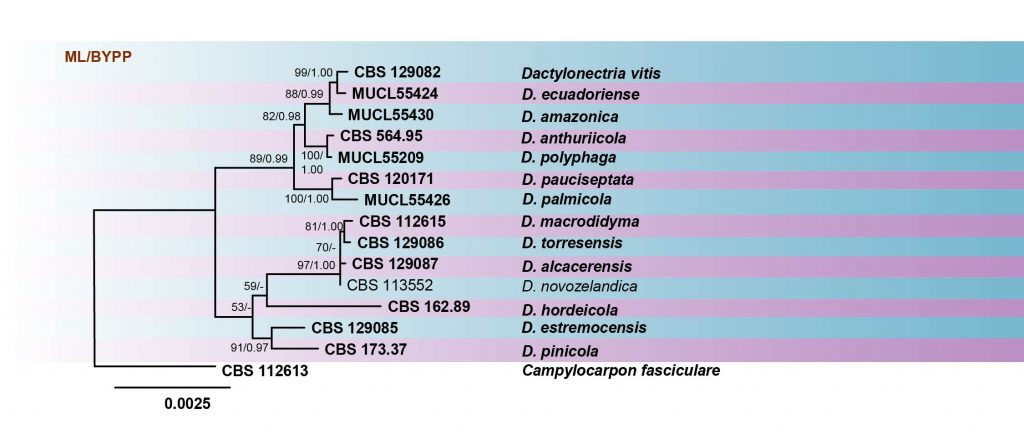
Lombard et al. (2014) re-evaluated genera with cylindrocarpon-like asexual morphs based on multi-gene phylogeny of ITS, LSU, TUB2 and tef1 genes. Gordillo and Decock (2018) analyzed His3 together with latter gene regions, for delimiting the species in Dactylonectria. Lombard et al. (2015) also supported the fact that Dactylonectria is monophyletic and distinct from Ilyonectria. In this study, we reconstruct the phylogeny of Dactylonectria based on analyses of a combined ITS, LSU, TUB2 and tef1 sequence data (Table 6, Fig. 12). The phylogenetic tree is updated with recently introduced Dactylonectria species and corresponds to previous studies (Lombard et al. 2014, 2015; Gordillo and Decock 2018).
Recommended genetic markers (genus level) – ITS, LSU, TUB2, tef1
Recommended genetic markers (species level) – TUB, tef1
The accepted number of species: 14 species
References: Cabral et al. 2012a, b; Halleen et al. 2004, Schroers et al. 2008; Lombard et al. 2014; Gordillo and Decock 2018 (morphology, phylogeny).
Table Details of the Dactylonectria isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate/Voucher no | ITS | LSU | TUB2 | tef1 |
| Campylocarpon fasciculare | CBS 112613* | AY677301 | HM364313 | AY677221 | JF735691 |
| Dactylonectria alcacerensis | CBS 129087* | JF735333 | KM231629 | AM419111 | JF735819 |
| D. anthuriicola | CBS 564.95* | JF735302 | KM515897 | JF735430 | JF735768 |
| D. amazonica | MUCL55433* | MF683707 | MF683727 | MF683644 | MF683665 |
| D. ecuadoriense | MUCL55424* | MF683704 | MF683724 | MF683641 | MF683662 |
| D. estremocencis | CBS 129085* | JF735320 | KM231630 | JF735448 | JF735806 |
| D. hordeicola | CBS 162.89* | AM419060 | KM515898 | AM419084 | JF735799 |
| D. macrodidyma | CBS 112615* | AY677290 | KM515900 | AY677233 | JF735836 |
| D. novozelandica | CBS 112608* | AY677288 | KM515901 | AY677235 | JF735821 |
| D. palmicola | MUCL55426* | MF683708 | MF683728 | MF683645 | MF683666 |
| D. pauciseptata | CBS 120171* | EF607089 | KM515903 | EF607066 | JF735776 |
| D. pinicola | CBS 173.37* | JF735319 | KM515905 | JF735447 | JF735803 |
| D. polyphaga | MUCL55209* | MF683689 | MF683710 | MF683626 | MF683647 |
| D. torresensis | CBS 129086* | JF735362 | KM231631 | JF735492 | JF735870 |
| D. vitis | CBS 129082* | JF735303 | KM515907 | JF735431 | JF735769 |
Fig. Phylogram generated from RAxML analysis based on combined sequences of ITS, LSU, TUB and tef1 sequences of all the accepted species of Dactylonectria. Related sequences were obtained from GenBank. Fifteen taxa are included in the analyses, which comprise 2460 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Campylocarpon fasciculare (CBS 112613). The tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of -6772.195394 is presented. The matrix had 261 distinct alignment patterns, with 0.96% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.230657, C = 0.279364, G = 0.252128, T = 0.237852; substitution rates AC = 1.388608, AG = 2.845402, AT = 2.389715, CG = 0.838197, CT = 7.220493, GT = 1.000000; gamma distribution shape parameter α = 0.650385. RAxML and maximum parsimony bootstrap support value ≥50% are shown respectively near the nodes. Bayesian posterior probabilities ≥0.95 (BYPP) indicated as thickened black branches. Ex-type strains are in bold.


No Comments