16 Oct Eutiarosporella
Eutiarosporella Crous, in Crous et al., Phytotaxa 202(2): 85 (2015)
Eutiarosporella was introduced by Crous et al. (2015) and is typified by Eutiarosporella tritici (B. Sutton & Marasas) Crous on Triticum aestivum from South Africa. The genus was named on account of its similarity to Tiarosporella Höhn. (Crous et al. 2006). Eutiarosporella species are coelomycetes that are saprobes or pathogens which occur in terrestrial habitats (Crous et al. 2015; Thynne et al. 2015; Li et al. 2016). Eutiarosporella species have been reported from Celtis africana N.L. Burm (Rosales), Triticum aestivum L. (Poales), Acacia karroo Hayne (Fabales) and Dactylis glomerata L. (Poales) (Thambugala et al. 2014; Crous et al. 2015). On wheat, it causes an economically important disease known as white grain disorder (Thynne et al. 2015). Several studies have reported this genus on woody hosts as a saprobe (Jami et al. 2012, 2014; Dissanayake et al. 2016).
Classification – Dothideomycetes, incertae sedis, Botryosphaeriales, Botryosphaeriaceae
Type species – Eutiarosporella tritici (B. Sutton & Marasas) Crous, in Crous et al., Phytotaxa 202(2):85 (2015)
Distribution – Worldwide
Disease symptoms – White grain disorder of wheat.
White grain disorder shrivels and discolors (white to light grey) wheat grain (Thynne et al. 2015). Affected grains are more brittle and can break during harvesting. Infected spikelets of greenheads may show bleaching appearance or grey discoloration. At first, the bleached florets may show blue-gray ‘highlights’. Rachis of affected heads and the upper peduncle may show a brownish discolouration (Thynne et al. 2015).
Even though species of this genus have been found to be associated with several hosts other than wheat, their diseases have not been described.
Hosts – Acacia karroo, Arrhenatherum elatius, Avenella flexuosa, Celtis africana, Dactylis glomerata, Triticum aestivum and Vachelloa karroo (Farr and Rossman 2019).
Morphological based identification and diversity
Eutiarosporella is characterized by hairy conidiomata with long necks, and holoblastic conidiogenesis, features which are clearly distinguishable from Tiarosporella (Höhnel 1919; Crous et al. 2015). This genus is morphologically similar to Marasasiomyces (long-necked, hairy conidiomata, and holoblastic conidiogenesis), except that it forms conidiomata in clusters, which are not found in Marasasiomyces (Crous et al. 2015). Li et al. (2016) reported the sexual morph of Eutiarosporella in E. dactylidis for the first time from Avenella flexuosa L. (Poales). The sexual morph comprises globose ascomata, with a central ostiole, a two-layered peridium, hyphae-like pseudoparaphyses and hyaline, aseptate, fusoid to ovoid ascospores, with a mucilaginous sheath (Thambugala et al. 2014).
Based on ITS and LSU sequence data, three species were initially included in this genus, E. africana (Jami, et al.) Crous, E. tritici (B. Sutton & Marasas) Crous and E. urbis-rosarum (Jami, et al.) Crous by Crous et al. (2015). Subsequently, E. darliae E. Thynne, et al., E. tritici-australis E. Thynne, et al. and E. dactylidis (Thambug., Camporesi & K.D. Hyde) Dissan., Camporesi & K.D. Hyde were accommodated in the genus (Crous et al. 2015; Thynne et al. 2015; Li et al. 2016), which now comprises seven species (Dissanayake et al. 2016; Wijayawardene et al. 2017).
Colony and conidial morphology are the primary characters to identify species within this genus. Colonies on nutrient-rich media (PDA or V8-OMA) grow rapidly (Thynne et al. 2015). However, we consider morphological characters alone are inadequate to identify species due to plasticity and overlapping of conidial dimensions. Therefore, the incorporation of molecular data together with morphology is recommended.
Molecular based identification and diversity
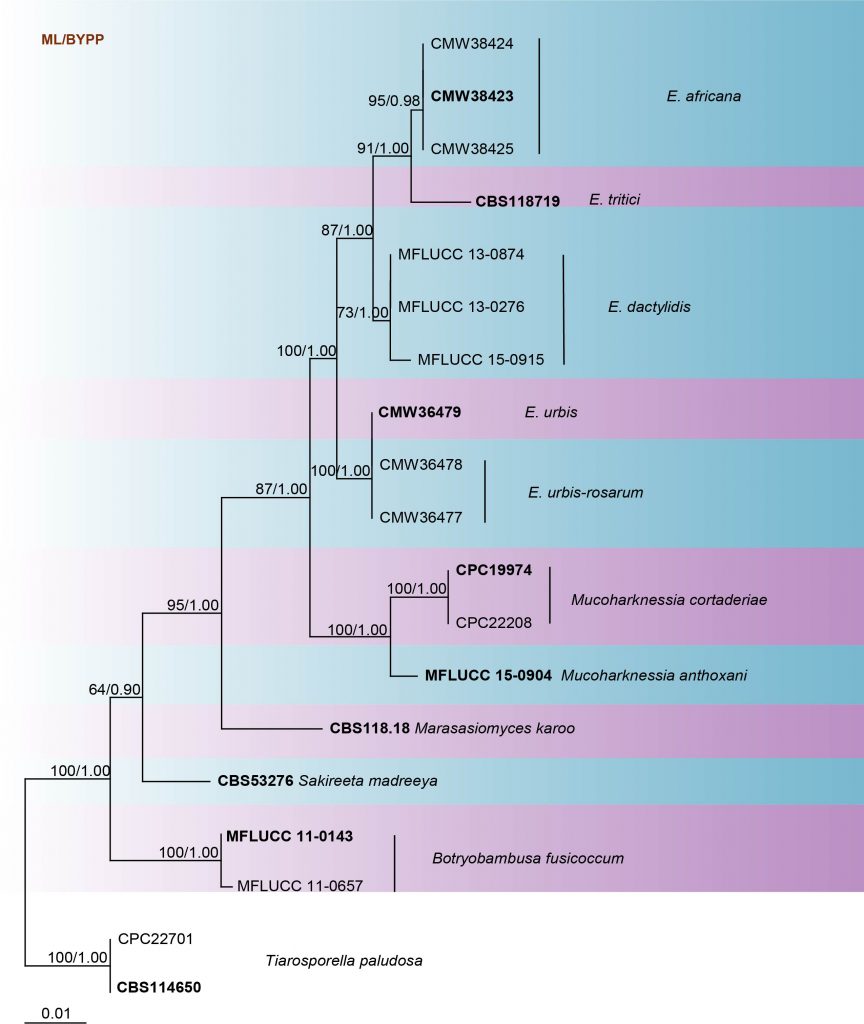
The taxonomy of Eutiarosporella is largely based on DNA sequence data to reveal the phylogenetic relationships between the species (Crous et al. 2015; Thynne et al. 2015; Dissanayake et al. 2016; Li et al. 2016). According to studies by Crous et al. (2015), Thynne et al. (2015) and Li et al. (2016), ITS and LSU are the most suitable loci for delineation of species within the genus. The phylogram generated with sequences available in GenBank including ex-epitype sequences is provided in Fig. 14. Our phylogenetic analyses are in accordance with previous studies by Crous et al. (2015), Thynne et al. (2015), Dissanayake et al. (2016) and Li et al. (2016).
Recommended genetic markers (Genus level) – LSU and SSU
Recommended genetic markers (Species level) – ITS and LSU
The accepted number of species: Seven species.
References: Crous et al. (2015), Thynne et al. (2015), Dissanayake et al. (2016), Li et al. 2016 (morphology, phylogeny).
Table Details of the Eutiarosporella isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate/Voucher no | LSU | ITS |
| Botryobambusa fusicoccum | MFLUCC 11-0657 | JX646810 | JX646793 |
| B. fusicoccum | MFLUCC 11-0143* | JX646809 | NR111793 |
| Eutiarosporella africana | CMW 38423* | KC769990 | KC769956 |
| E. dactylidis | MFLUCC 15-0915 | KU246380 | NR148093 |
| Eutiarosporella dactylidis | MFLUCC 13-0874 | KM978948 | KM978945 |
| E. dactylidis | MFLUCC 13-0276* | KM978949 | KM978944 |
| E. urbis | CMW 36477* | JQ239420 | NR111705 |
| E. urbis-rosarum | CMW 36479 | JQ239422 | JQ239409 |
| E. urbis-rosarum | CMW 36478 | JQ239421 | JQ239408 |
| Marasasiomyces karoo | CBS 1187.18* | DQ377939 | KF531828 |
| Mucoharknessia anthoxanthi | MFLUCC 15-0904* | KU246379 | NR148092 |
| M. cortaderiae | CPC 22208 | KM108402 | KM108375 |
| M. cortaderiae | CPC 19974* | KM108401 | NR148075 |
| Sakireeta madreeya | CBS 532.76* | DQ377940 | KC769960 |
| Tiarosporella africana | CMW 38425 | KC769992 | KC769958 |
| T. africana | CMW 38424 | KC769991 | KC769957 |
| T. paludosa | CPC 22701 | KM108404 | NR132907 |
| T. paludosa | CBS 114650* | KM108403 | KM108377 |
| T. tritici | CBS 118719* | DQ377941 | KC769961 |
Fig. Phylogram generated from maximum likelihood analysis based on combined LSU and ITS sequence data retrieved from GenBank. The tree is rooted in Tiarosporella paludosa (CPC 22701 and CBS 114650). Tree topology of the ML analysis was similar to the Bayesian analysis. The best scoring RAxML tree with a final likelihood value of -7055.996836 is presented. The matrix had 380 distinct alignment patterns, with 40.12% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.229604, C = 0.264209, G = 0.282595, T = 0.223593; substitution rates AC = 1.357965, AG = 1.612491, AT = 0.913118, CG = 2.194420, CT = 5.121479, GT = 1.000000; gamma distribution shape parameter α = 0.137391. Maximum likelihood bootstrap support values greater than 60% are indicated above the nodes. Ex-type (ex-epitype) and voucher strains are in bold.

No Comments