16 Oct Macrophomina
Macrophomina Petr., Annls mycol. 21: 314 (1923)
Species of Macrophomina are mostly pathogens that cause damping-off, seedling blight, collar rot, stem rot, charcoal rot, basal stem rot and root rot in many plant species (Arora et al. 2001; Pal et al. 2001; Gupta et al. 2002; Sarr et al. 2014; Wijayawardene et al. 2017). The type species, Macrophomina phaseolina (Tassi) Goid., is a seed-borne polyphagous pathogen that affects more than 500 crop and non-crop species, including economically important crops, such as soybean, sunflower, common bean, peanut, corn, sorghum, cowpea and cotton (Gupta et al. 2002; Ndiaye et al. 2010; Sarr et al. 2014).
Classification – Dothideomycetes, incertae sedis, Botryosphaeriales, Botryosphaeriaceae
Type species – Macrophomina phaseolina (Tassi) Goid., Annali Sper. agr., N.S. 1(3): 457 (1947)
Distribution – Worldwide
Disease symptoms – Charcoal rot, collar rot, damping-off, root rot, seedling blight, stem rot, wilt
Seedling damage can occur when infected seeds are planted. Infected plants may produce slightly smaller leaflets than healthy plants and have reduced vigor. As the disease advances, leaflets turn yellow, wilt and turn brown (Adorada et al. 2018). A grey/silver discoloration can be observed in the roots and lower stem when the plants split open (Romero et al. 2017; Koehler and Shew 2018; Meena et al. 2018). In charcoal rot, the abundant production of minute black sclerotia by the fungus causes the rotted tissues to become blackened. Infections on soybean lead to early maturation and incomplete pod filling (ElAraby et al. 2003; Yang and Navi 2005; Sarr et al. 2014). In peanut, it causes seed and seedling rots, wilt, root and stem rots, leaf spot and rotting of developing pods and seeds (Gupta et al. 2002; Deshwal et al. 2003).
Hosts – This soil-borne fungus can infect more than 500 agricultural crops and weed species including, Fragaria, Glycine, Helianthus, Sorghum, and Zea.
Morphological based identification and diversity
Eight species names are recorded in Index Fungorum (2019), however, sequences are available for only two species Macrophomina phaseolina and M. pseudophaseolina (Sarr et al. 2014). Morphological characteristics of M. phaseolina are mostly similar to M. pseudophaseolina, except that conidia of the latter are shorter.
Colony and conidial morphology are the primary characters used to identify species within this genus (Ellis 1971, 1976; Simmons 1992). However, the connectivity of sexual and asexual morphs is not proven, as no sexual morph has been obtained from nature or culture (Crous et al. 2006; Wijayawardene et al. 2016, 2017). According to the morphological identifications, Macrophomina phaseolina has conidia with apical mucoid appendages as found in Tiarosporella (Sutton and Marasas 1976). Nevertheless, it can be distinguished from Tiarosporella in having conidia with apical mucoid appendages, per currently proliferating conidiogenous cells and dark brown (at maturity) conidia (Crous et al. 2006; Phillips et al. 2013). Morphologically M. phaseolina is similar to M. pseudophaseolina, except that conidia of the latter are shorter.
Molecular based identification and diversity
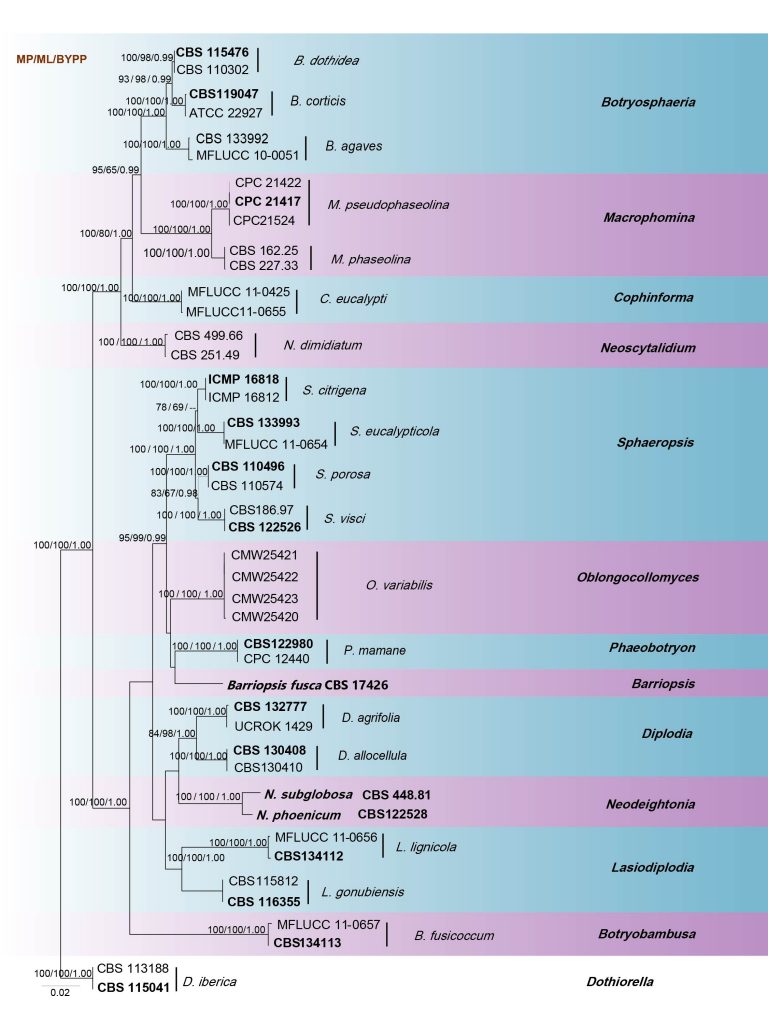
Phillips et al. (2013) suggested that phylogenetic analysis of combined SSU, LSU, ITS, tef1 and TUB2 genes provide better resolution. Sarr et al. (2014) used ITS, tef1, ACT, CAL and TUB2 sequence data representing a large sample of Macrophomina isolates from many hosts. According to the multi-gene analysis of SSU, LSU, ITS, tef1 and TUB2 genes in this study (Fig. 16), the two species cluster in a well-supported clade with high bootstrap values (100% ML, 1.00 BYPP). The overall topology of our phylogeny tree is similar to previous studies.
Recommended genetic markers (genus level) – LSU and SSU
Recommended genetic markers (species level) – ITS, tef1, ACT, CAL and TUB2
The accepted number of species: There are eight epithets in Index Fungorum (2019) However, two species have molecular data.
References: Crous et al. 2006; Phillips et al. 2013; Sarr et al. 2014, Wijayawardene et al. 2016 (morphology, phylogeny).
Table. Details of Macrophomina and Sphaeropsis isolates used in the phylogenetic analyses. Ex-type (or ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher no | SSU | ITS | LSU | tef1 | TUB2 |
| Barriopsis fusca | CBS 174.26* | EU673182 | EU673330 | DQ377857 | EU673296 | EU673109 |
| Botryobambusa fusicoccum | CBS 134113* | JX646826 | JX646792 | JX646809 | JX646857 | N/A |
| MFLUCC 11-0657 | JX646827 | JX646793 | JX646810 | JX646858 | N/A | |
| Botryosphaeria agaves | CBS 133992* | JX646825 | JX646791 | JX646808 | JX646856 | JX646841 |
| MFLUCC 10-0051 | JX646824 | JX646790 | JX646807 | JX646855 | JX646840 | |
| B. corticis | CBS 119047* | EU673175 | DQ299245 | EU673244 | EU017539 | EU673107 |
| ATCC 22927 | EU673176 | DQ299247 | EU673245 | EU673291 | EU673108 | |
| B. dothidea | CBS 115476 * | EU673173 | AY236949 | AY928047 | AY236898 | AY236927 |
| CBS 110302 | EU673174 | AY259092 | EU673243 | AY573218 | EU673106 | |
| Cophinforma atrovirens | MFLUCC 11-0425* | JX646833 | JX646800 | JX646817 | JX646865 | JX646848 |
| MFLUCC 11-0655 | JX646834 | JX646801 | JX646818 | JX646866 | JX646849 | |
| Dothiorella iberica | CBS 115041* | EU673155 | AY573202 | AY928053 | AY573222 | EU673096 |
| CBS 113188 | EU673156 | AY573198 | EU673230 | EU673278 | EU673097 | |
| Diplodia allocellula | CBS 130408* | N/A | JQ239397 | JQ239410 | JQ239384 | JQ239378 |
| CBS 130410 | N/A | JQ239399 | JQ239412 | JQ239386 | JQ239380 | |
| D. agrifolia | CBS 132777* | N/A | JN693507 | N/A | JQ517317 | JQ411459 |
| UCROK 1429 | N/A | JQ411412 | N/A | JQ512121 | JQ411443 | |
| Lasiodiplodia gonubiensis | CBS 115812* | EU673193 | AY639595 | DQ377902 | DQ103566 | DQ458860 |
| CBS 116355 | EU673194 | AY639594 | EU673252 | DQ103567 | EU673126 | |
| L. lignicola | CBS 134112* | JX646830 | JX646797 | JX646814 | JX646862 | JX646845 |
| MFLUCC 11-0656 | JX646831 | JX646798 | JX646815 | JX646863 | JX646846 | |
| Neoscytalidium hyalinum | CBS 499.66 | KF531818 | KF531820 | DQ377925 | KF531798 | KF531800 |
| CBS 251.49 | KF531817 | KF531819 | DQ377923 | KF531797 | KF531799 | |
| Neodeightonia subglobosa | CBS 448.81* | EU673202 | EU673337 | DQ377866 | EU673306 | EU673137 |
| N. phoenicum | CBS 122528* | EU673205 | EU673340 | EU673261 | EU673309 | EU673116 |
| Macrophomina phaseolina | CBS 227.33 | KF531823 | KF531825 | DQ377906 | KF531804 | KF531806 |
| CBS 162.25 | KF531824 | KF531826 | DQ377905 | KF951996 | KF531805 | |
| M. pseudophaseolina | CPC 21422 | N/A | KF951792 | N/A | KF952154 | KF952234 |
| CPC 21417* | N/A | KF951791 | N/A | KF952153 | KF952233 | |
| CPC 21524 | N/A | KF951799 | N/A | KF952161 | KF952240 | |
| Phaeobotryon mamane | CBS 122980* | EU673184 | EU673332 | EU673248 | EU673298 | EU673121 |
| CPC 12442 | EU673185 | EU673333 | DQ377899 | EU673299 | EU673124 | |
| Oblongocollomyces variabilis | CMW 25420 | N/A | EU101313 | N/A | EU101358 | N/A |
| CMW 25421, CBS 121775 | N/A | EU101314 | N/A | EU101359 | N/A | |
| CMW 25422, CBS 121776 | N/A | EU101326 | N/A | EU101371 | N/A | |
| CMW 25423 | N/A | EU101327 | N/A | EU101372 | N/A | |
| Sphaeropsis citrigena | ICMP 16812* | EU673180 | EU673328 | EU673246 | EU673294 | EU673140 |
| ICMP 16818 | EU673181 | EU673329 | EU673247 | EU673295 | EU673141 | |
| S. eucalypticola | CBS 133993* | JX646835 | JX646802 | JX646819 | JX646867 | JX646850 |
| MFLUCC 11-0654 | JX646836 | JX646803 | JX646820 | JX646868 | JX646851 | |
| S. porosa | CBS 110496* | EU673179 | AY343379 | DQ377894 | AY343340 | EU673130 |
| CBS 110574 | N/A | AY343378 | N/A | AY343339 | N/A | |
| S. visci | CBS 122526 * | N/A | EU673324 | N/A | EU673292 | N/A |
| CBS 186.97 | EU673178 | EU673325 | DQ377868 | EU673293 | EU673128 |
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU, SSU, tef1 and TUB2 sequences. Related sequences were obtained from GenBank. Forty-four strains are included in the analyses, which comprise 3477 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Dothiorella iberica (CBS 113188 and CBS 115041). The tree topology of the ML analysis was similar to the MP and BI. The best scoring RAxML tree with a final likelihood value of -12764.659013 is presented. The matrix had 898 distinct alignment patterns, with 28.31% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.238049, C = 0.253522, G = 0.272022, T = 0.236408; substitution rates AC = 1.121500, AG = 2.393284, AT = 1.053637, CG = 1.711098, CT = 4.682724, GT = 1.000000; gamma distribution shape parameter α = 0.545763. The maximum parsimonious dataset consisted of constant 2820, 575 parsimony-informative and 82 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 1490 steps (CI = 0.626, RI = 0.861, RC = 0.539, HI = 0.374) in the first tree. RAxML, maximum parsimony bootstrap support values ≥65% and Bayesian posterior probabilities ≥0.95 (BYPP) are shown respectively near the nodes. Ex-type strains are in bold.

No Comments