16 Oct Neopestalotiopsis
Neopestalotiopsis Maharachch., K.D. Hyde & Crous (2014), in Maharachchikumbura et al., Stud. Mycol. 79:147 (2014)
Neopestalotiopsis is an important plant pathogenic, saprobic and endophytic genus commonly present in tropical and subtropical ecosystems. The genus was introduced by Maharachchikumbura et al. (2014b). Species of Neopestalotiopsis are appendage-bearing asexual coelomycetes in the family Sporocadaceae (Jayawardena et al. 2016).
Classification – Sordariomycetes, Xylariomycetidae, Amphisphaeriales, Sporocadaceae
Type species – Neopestalotiopsis protearum (Crous & L. Swart) Maharachch. et al., in Maharachchikumbura et al., Stud. Mycol. 79:147 (2014)
Distribution – Worldwide
Disease symptoms – Canker, dieback, fruit rots, leaf spot
Pathogenic Neopestalotiopsis are recorded in post-harvest fruit rots of grapes, trunk diseases in grapevine in China, India and France, leaf spot disease of grapevine in China and leaf blights in many plant species worldwide (Hyde et al. 2014; Jayawardena et al. 2015, 2016; Maharachchikumbura et al. 2017).
Neopestalotiopsis species infect a variety of grapevine cultivars, causing diseases including grapevine dieback, fruit rot, postharvest disease, and severe defoliation. Initial symptoms of fruit rot disease are mostly observed at the splits between the pedicel and the berry and at the wounds of the fruits and severely infected fruits become rotten and separate completely from the pedicel (Jayawardene et al. 2015). Neopestalotiopsis asiatica and N. javaensis are associated with grapevine trunk disease (Maharachchikumbura et al. 2017). Grapevine trunk diseases reduce the yield and quality of grapes, even leading to partial or total death of individual plants.
Neopestalotiopsis clavispora and N. surinamensis cause guava scab (Solarte et al. 2018). Neopestalotiopsis ellipsospora causes leaf spots on sweet potatoes (Maharachchikumbura et al. 2016). Neopestalotiopsis clavispora causes crown and root rot of strawberry worldwide while N. iranensis infects leaves and fruits of strawberry (Ayoubi and Soleimani 2016), with the pathogen initially developing circular, black, and slightly sunken spots that expand outwards on the surface. Droplets of spores are scattered over the white aerial mycelial area and later cause soft decay of the fruit flesh (Ayoubi and Soleimani 2016).
Canker and dieback on blueberry in Chile and Uruguay are also caused by N. clavispora (Espinoza et al. 2008; González et al. 2012; Chamorro et al. 2016). Neopestalotiopsis samarangensis has been described from wax apple fruit rot in Thailand (Maharachchikumbura et al. 2013). In fruit rots, the initial symptom is small, circular, black, slightly sunken spots on fruits. Later, the spots enlarged rapidly, become sunken and result in a soft decay of the fruit flesh (Maharachchikumbura et al. 2013).
Hosts – Species of Fragaria × ananassa, Ipomoea, Malus, Psidium, Vaccinium and Vitis
Morphological based identification and diversity
Neopestalotiopsis species can be differentiated using morphology and molecular phylogeny (Maharachchikumbura et al. 2014b). There are 36 species epithets are listed in Index Fungorum (2019). Neopestalotiopsis species differ from Pestalotiopsis and Pseudopestalotiopsis in having somewhat versicolorous median cells (Maharachchikumbura et al. 2014b) whereas both Pestalotiopsis and Pseudopestalotiopsis have concolorous median cells (Maharachchikumbura et al. 2014b) as well as its conidiophores which are indistinct and often reduced to conidiogenous cells (Maharachchikumbura et al. 2014b).
Conidial morphology is widely used in taxonomy in pestalotioid fungi (Steyaert 1949; Guba 1961; Nag Raj 1993; Maharachchikumbura et al. 2012, 2014b). Species delimitation based on morphological characters is limited as these characters are plastic and vary between hosts and environments (Maharachchikumbura et al. 2011, 2016). Therefore, phylogenetic species recognition is an effective method to identify different pestalotioid species (Maharachchikumbura et al. 2016).
Molecular based identification and diversity
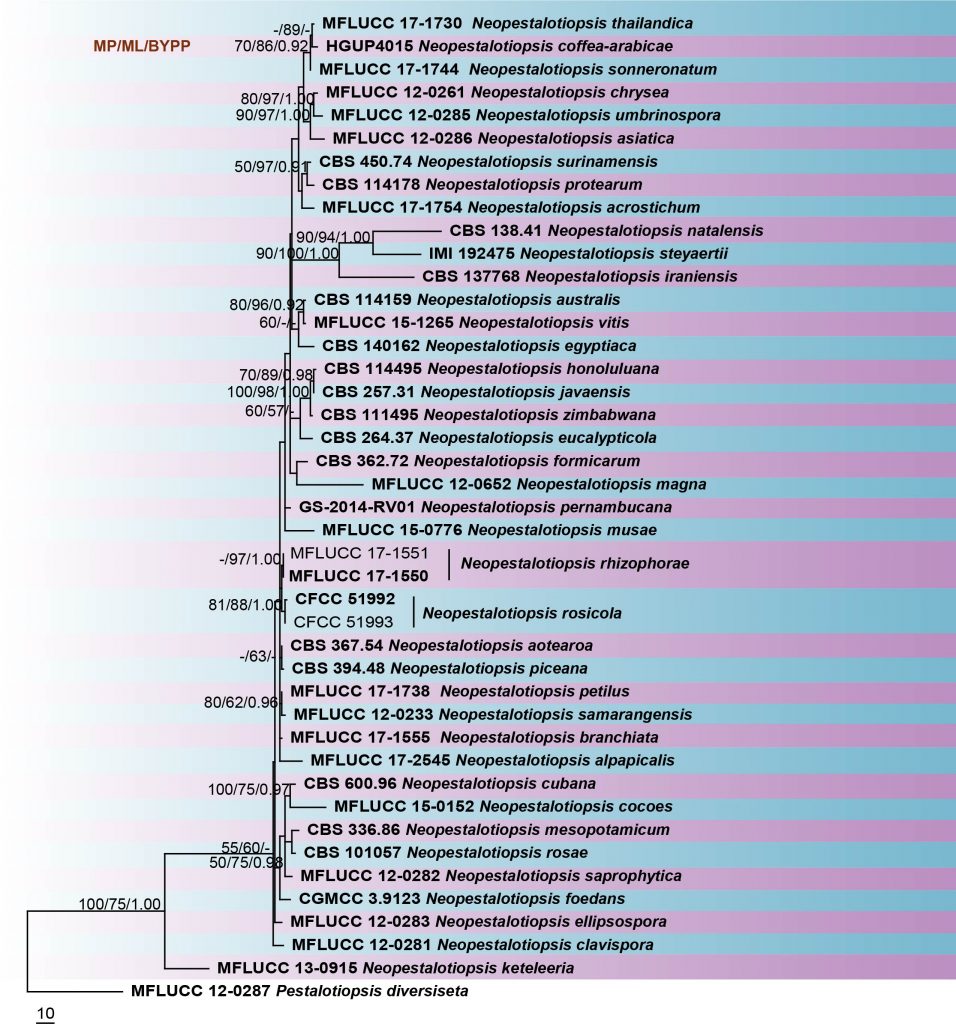
Neopestalotiopsis species can be roughly separated from Pestalotiopsis and Pseudopestalotiopsis based on the total number of base pairs in the ITS region (Maharachchikumbura et al. 2014b). However, the use of ITS sequences alone does not resolve Neopestalotiopsis species (Maharachchikumbura et al. 2012). Therefore, Maharachchikumbura et al. (2014b) suggested using combined ITS, TUB2 and tef1 genes to provide a better resolution in phylogenetic analyses. This study reconstructs the phylogeny of Neopestalotiopsis based on combined ITS, TUB2 and tef1 sequence data (Fig 18) and reveals similar phylogenetic relationships to previous studies by Maharachchikumbura et al. (2014b, 2016).
Recommended genetic markers (genus level) – LSU
Recommended genetic markers (species level) – ITS, TUB2 and tef1
The accepted number of species: 41 species.
References: Maharachchukumbura 2012, 2014b (morphology, phylogeny); Maharachchukumbura 2016 (morphology, phylogeny); Jayawardena et al. 2015, 2016 (morphology, phylogeny, pathogenicity)
Table. Details of the Neopestalotiopsis isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate no | ITS | TUB2 | tef1 |
| Neopestalotiopsis acrostichum | MFLUCC 17-1754* | MK764272 | MK764338 | MK764316 |
| N. alapicalis | MFLUCC 17-2544* | MK357772 | MK463545 | MK463547 |
| N. aotearoa | MFLUCC 17-1754 | MK764272 | MK764338 | MK764316 |
| N. asiatica | MFLUCC12-0286* | JX398983 | JX399018 | JX399049 |
| N. australis | CBS 114159* | KM199348 | KM199432 | KM199537 |
| N. brachiata | MFLUCC 17-1555* | MK764274 | MK764340 | MK764318 |
| N. chrysea | MFLUCC12-0261* | JX398985 | JX399020 | JX399051 |
| N. clavispora | MFLUCC 12-0281* | JX398979 | JX399014 | JX399045 |
| N. cocoes | MFLUCC 15-0152* | KX789687 | – | KX789689 |
| N. coffeae-arabicae | HGUP 4019* | KF412647 | – | – |
| N. cubana | CBS 600 96* | KM199347 | KM199438 | KM199521 |
| N. egyptiaca | CBS 140162* | KP943747 | KP943746 | KP943748 |
| N. ellipsospora | MFLUCC 12-0283* | JX398980 | JX399016 | JX399047 |
| N. eucalypticola | CBS 264 37* | KM199376 | KM199431 | KM199551 |
| N. foedans | CGMCC3 9123* | JX398987 | JX399022 | JX399053 |
| N. formicarum | CBS 362 72* | KM199358 | KM199455 | KM199517 |
| N. honoluluana | CBS 114495* | KM199364 | KM199457 | KM199548 |
| N. iraniensis | CBS 137768* | KM074048 | KM074057 | KM074051 |
| N. javaensis | CBS 257 31* | KM199357 | KM199437 | KM199543 |
| N. keteleeria | MFLUCC 13-0915* | KJ503820 | KJ503821 | KJ503822 |
| N. macadamiae | BRIP 63738B* | KX186604 | KX186654 | KX186627 |
| N. magna | MFLUCC12-652* | KF582795 | KF582793 | KF582791 |
| N. mesopotamica | CBS 336 86* | KM199362 | KM199441 | KM199555 |
| N. musae | MFLUCC 15-0776* | KX789683 | KX789686 | KX789685 |
| N. natalensis | CBS 138 41* | KM199377 | KM199466 | KM199552 |
| N. pernambucana | GS-2014 strain RV01* | KJ792466 | – | KU306739 |
| N. petila | MFLUCC 17-1738* | MK764275 | MK764341 | MK764319 |
| N. piceana | CBS 394 48* | KM199368 | KM199453 | KM199527 |
| N. protearum | CBS 114178* | JN712498 | KM199463 | KM199542 |
| N. rhisophorae | MFLUCC 17-1550* | MK764277 | MK764343 | MK764321 |
| N. rosae | CBS 101057* | KM199359 | KM199429 | KM199523 |
| N. rosicola | CFCC 51992 | KY885239 | KY885245 | KY885243 |
| N. samarangensis | MFLUCC 12-0233* | JQ968609 | JQ968610 | JQ968611 |
| N. saprophytica | MFLUCC 12-0282* | KM199345 | KM199433 | KM199538 |
| N. sonneratae | MLFUCC 17-1745* | MK764279 | MK264345 | MK264323 |
| N. steyaertii | IMI192475* | KF582796 | KF582794 | KF582792 |
| N. surinamensis | CBS 450.74* | KM199351 | KM199465 | KM199518 |
| N. thailandica | MFLUCC 17-1730* | MK764281 | MK764347 | MK754325 |
| N. umbrinospora | MFLUCC 12-0285* | JX398984 | JX399019 | JX399050 |
| N. vitis | MFLUCC 15-1265* | KU140694 | KU140685 | KU140676 |
| N. zimbabwana | CBS 111495* | – | KM199456 | KM199545 |
Fig. Phylogram generated from maximum likelihood analysis based on combined ITS, TUB2 and tef1 sequence data of Neopestalotiopsis species. Related sequences were obtained from GenBank. Forty-three strains are included in the combined sequence analyses, which comprise 1391 characters with gaps. Pestalotiopsis diversiseta (MFLUCC 12-0287) is used as the outgroup taxa. The best scoring RAxML tree with a final likelihood value of -5457.035085 is presented. The matrix had 409 distinct alignment patterns, with 6.30% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.231067, C = 0.270889, G= 0.213946, T = 0.284098; substitution rates AC = 0.847461, AG = 2.876343, AT = 1.282349, CG = 0.723831, CT = 3.850003, GT = 1.000000; gamma distribution shape parameter α = 0.235476. The maximum parsimonious dataset consisted of 1026 constant, 177 parsimony-informative and 188 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of ten equally most parsimonious trees with a length of 650 steps (CI = 0.688, RI = 0.609, RC = 0.419, HI = 0.312) in the first tree. RAxML and maximum parsimony bootstrap support value ≥50% are shown respectively near the nodes. Ex-type strains are in bold.

No Comments