23 Oct Armillaria
Armillaria (Fr.) Staude, Schwämme Mitteldeutschl. 28: xxviii, 130 (1857)
Background
Armillaria is a plant pathogenic genus in the phylum Basidiomycota, family Physalacriaceae (He et al. 2019), collectively referred to as shoestring root-rot fungi or honey mushrooms. Armillaria can cause root-rot disease in a wide variety of woody hosts worldwide. Armillaria has undergone significant revision in the past 20 years. The genus once accommodated any white-spored agaric with broadly attached gills and an annulus (Volk et al. 1996). Armillaria mellea is the type species. Most Armillaria species have the potential to infect healthy and stressed trees, they differ in their pathogenicity to their hosts and under certain circumstances, they behave as obligate saprobes. Most Armillaria species are facultative necrotrophs causing root and butt rot on a broad range of woody plants affecting a variety of forest, shade, ornamental and orchard trees and shrubs. Some Armillaria species cause significant economic losses to forest trees and in nursery plantations. Armillaria root disease is found in many temperate and tropical forests throughout the world. This fungus spreads mainly through the interaction of tree roots. As saprotrophs, Armillaria species are important wood decomposers that contribute to nutrient cycling in forest ecosystems. As pathogens, they infect and eventually kill susceptible trees, which impacts forest structure, composition and succession. Trees that are used for fibre or lumber production, as well as trees located in recreation sites, are affected by these diseases. Such Armillaria infections may cause yield reduction and tree mortality in silvicultural and agricultural tree plantations and provoke economic losses.
Armillaria species are expected to become more aggressive during drought and thus enhance root rot (La Porta et al. 2008; Kolb et al. 2016; Kubiak et al. 2017). The incidence of Armillaria related root disease is likely to increase as temperatures increase and precipitation decreases due to climate change (Sturrock et al. 2011). Whilst the ability of the pathogen to sporulate, spread and infect is affected by temperature and moisture, factors that stress host trees directly may be just as critical to a successful invasion of host tissues. It seems likely that the disease will become more severe in the future, wherever Armillaria susceptible tree species are subjected to increased levels of climate stress (Klopfenstein et al. 2009). Currently, Armillaria root disease causes large growth/volume losses (e.g., 16–55%) in areas of western and North America (Filip and Goheen 1984; Cruickshank Morrison et al. 2011; Lockman and Kearns 2016). Armillaria root disease is typically more severe in trees that are maladapted to climate-induced stress (Ayres and Lombardero 2000; Kliejunas et al. 2009; Sturrock et al. 2011). Thus, it is likely that climate change will further exacerbate damage from Armillaria root disease, which can further predispose trees to beetle attack (e.g. Hertert et al. 1975; Tkacz and Schmitz 1986; Goheen and Hansen 1993).
Armillaria mellea is an edible species that has long been used as a Traditional Chinese Medicine. Some of Armillaria species are is believed to be able to improve health and prevent various diseases, such as insomnia, pain, and neurasthenia. Extracts of A. mellea exhibit anti-oxidative, anti-inflammatory and immune-modulatory activities. Armillaria mellea can also induce maturation of human dendritic cells. The chemical constituents isolated from A. mellea include sesquiterpenoids, steroids, triterpenoids, adenosine and resin acids. Armillariol C is a furan-based natural product isolated from Armillaria species. A xylosyl 1,3-galactofucan (AMPS-III) was isolated and identified as a novel anti-inflammatory agent from this species.
Classification – Basidiomycota, Agaricomycotina, Agaricomycetes, Agaricomycetidae, Agaricales, Physalacriaceae (He et al. 2019)
Type species – Armillaria mellea (Vahl) P. Kumm.
Distribution – Worldwide, mostly in temperate areas (northern and southern hemisphere) and some in tropical areas.
Disease symptoms – Armillaria root disease, shoestring root rot
Symptoms caused by this fungus can be categorized into two categories:
Crown symptoms—branch dieback, crown thinning, chlorosis, reddening of foliage or heavier than normal production of cones.
Basal symptoms —the fungus can grow up from the roots in the inner bark in some tree species and causes basal cankers above the infected roots. Resinosis (exudation of resin) can be observed in resinous conifers. In some plants, decayed roots or decay in the inner wood of stem bases can be observed. Species cause a white rot of wood. In white rot, wood often has a bleached, whitish appearance and are spongy or stringy, and maybe wet. Black lines called “zone lines” are usually seen in the decayed wood. These lines are curved planes in the wood, sometimes called “pseudosclerotial plates”, composed of thickened, dark fungal cells. They may play a role in the protection of Armillaria from unfavourable conditions or other fungi that attempt to invade its territory, including other individuals of the same species. Actively decaying wood may be luminescent, producing a faint glow in the dark (Baumgartner et al. 2002; Worrall 2004; Klopfenstein 2009).
There are three major signs of Armillaria root disease in the field.
Mycelial fans can always be seen in infected and recently killed trees. These are white mats of fungal mycelium between the inner bark and wood that are generally substantial and have a mushroom odour.
Rhizomorphs are commonly associated with infection and are often attached to infected roots, but they may also be attached to the surface of uninfected roots. Depending on the species these may be few, small, fragile, hard to find or abundant and robust. Rhizomorphs can be cylindrical in soil or flattened under bark, reddish-brown to black branched and have a cream-coloured tip when actively growing (Guillaumin and Legrand 2013).
Mushrooms that have honey-brown caps can be seen in clusters near or on the base of trees.
Hosts – Many angiosperms and gymnosperms (especially conifers) in native, planted forests, orchards and vineyards (Farr and Rossman 2020).
Pathogen biology, disease cycle and epidemiology
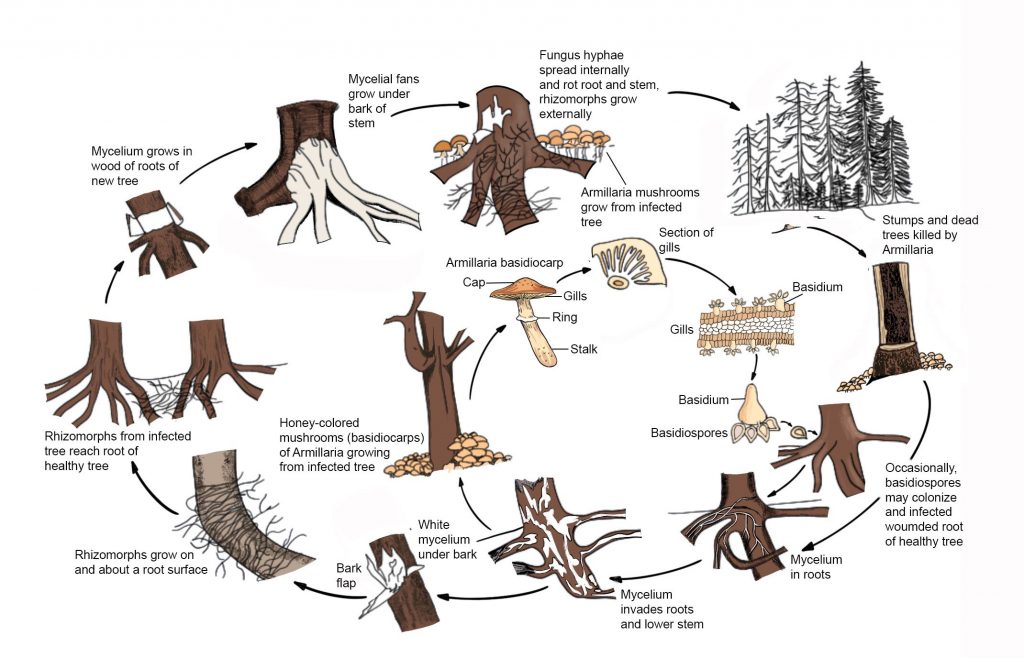
Sexual reproduction results in the diploid mycelium. Such a mycelium is the dominant phase that is found growing in wood, growing through the soil as rhizomorphs, and killing trees. Armillaria species can be dispersed through airborne sexual basidiospores which will establish a new infection center. These taxa do not reproduce asexually but disperse by growing mycelium which is the most common source of infection, through root contacts or root grafts or by growing through the soil as rhizomorphs. Mycelium in colonized roots and the rhizomorphs produced serve as the most common mode of infection and may survive for up to 50 years or more in stumps, depending on the climate, size of the stump, and other factors (Baumgartner et al. 2002; Worrall 2004; Klopfenstein 2009).
Morphology-based identification and diversity
Armillaria has included only white-spored wood-inhabiting agarics with broadly attached to decurrent gills and macroscopic black to reddish-brown rhizomorphs. Armillaria basidiomes are easily recognized by their caespitose habit, annulus and honey colour. It is, however, extremely difficult to identify some species due to the lack of morphological apomorphies (Watling et al. 1991; Pegler 2000). Besides, basidiomata are often not available to differentiate species, which further complicates the taxonomy of Armillaria (Harrington and Wingfield 1995). In this regard, Armillaria provides a clear example of where a phylogenetic approach can contribute significantly to its taxonomy. Until the late 1970s, Armillaria mellea was considered by most researchers to be a polymorphic species with a wide host range and distribution. Herink (1973), among others, suspected that this single species might be a species complex. However, since the morphology of basidiomata is difficult to study because of overlapping and inconsistent traditionally used morphological characters, other avenues of research were pursued. Hintikka (1973) developed a technique that allowed the determination of mating types in Armillaria. Using a modification of this method, Korhonen (1978a) was able to distinguish five European biological species. The cumbersome nature of the mating-type method of species identification prompted a search for other techniques for identifying collections. They were able to separate all North American species (NABS) of Armillaria except for A. calvescens and A. gallica, which are apparently very closely related (Anderson and Stasovski1992). Ten species of Armillaria in North America have been confirmed from multiple studies utilizing a combination of morphological, biological and phylogenetic species concepts (Anderson and Ullrich 1979; Anderson and Stasovski 1992; Burdsall and Volk 1993; Kim et al. 2006; Ross-Davis et al. 2012). Before, A. mellea shows great variability in morphology and hosts. These species were first separated using interfertility tests using cultures of Armillaria haploid tester strains and morphology. Now, A. mellea is considered as an independent species, with two North American biological species (Bérubé and Dessureault 1989; Volk et al. 1996).
Fig. 1 Disease cycle of Armillaria mellea (redrawn from Agrios 2005)
Molecular-based identification and diversity
Problems surrounding the identification of Armillaria have led to important advances in developing robust but rapid DNA techniques. Such techniques have initially included DNA-base composition (Jahnke et al. 1987) DNA-DNA hybridization (Miller et al. 1994), sequence analyses of the IGS-1(Anderson and Stasovski 1992) and ITS (Coetzee et al. 2001a,b), RFLPs without PCR (Smith and Anderson 1989) and RFLPs of IGS-1 amplicons (Harrington and Wingfield 1995). Although several of these techniques might pose some problems (Pérez‐Sierra et al. 2000), by their relative simplicity they have gradually replaced traditional, morphological methods.
The amount of DNA sequence data on Armillaria species has increased substantially since the first publication on the phylogeny of the genus in the northern hemisphere (Anderson and Stasovski 1992). As with many other fungal genera, the focus of such studies initially was set on species of Europe and North America (Chillali et al. 1998; Coetzee et al. 2000b). Later, substantial datasets for species in Africa, Australasia and southeast Asia have become available (Terashima et al. 1998; Coetzee et al 2000a; 2001a). At present, ITS, IGS-1 and tef1 sequences are available in GenBank for the best-known species of Armillaria. However, there are disjunctions in data sets and relatively little is known about species from Indo-Malaysia and South America. Armillaria fruiting bodies are produced seasonally and not every year; they are, therefore, often not available during fieldwork (Kile et al. 1991).
Identification using the biological species concept with species identification based on sexual compatibility tests (Korhonen 1978a) has been examined for its utility by some mycologists, but its application was soon abandoned. This was because of complications due to the absence of known tester strains, lack of haploid strains, ambiguous mating interactions and degeneracy of cultures. For these reasons, DNA-based molecular techniques have finally been preferred in Armillaria taxonomy, either complementing other methods or on their own. The techniques utilized for the taxonomy of Armillaria species include comparisons of RFLPs (Harrington and Wingfield 1995), AFLPs (Pérez-Sierra et al. 2004), and the use of sequences from the ITS, IGS-1 and tef1 gene in phylogenetic studies (Coetzee et al. 2000b, 2001a; Maphosa et al. 2006; Kim et al. 2006). Phylogenetic methods have made it possible to differentiate the lineages of the genus in southern Argentina (Pildain et al. 2009). Lineages I and II grouped with A. novae-zelandiae and A. luteobubalina, respectively, while Lineages III and IV represented unique taxa that were closely related to A. hinnulea, Armillaria 4th species from New Zealand (established by Coetzee et al. 2001) and Armillaria Group III from Kenya (Mwenje et al. 2006). Modern approaches to identification of Armillaria species are mostly based on the analyses of DNA sequences. The present study reconstructs the phylogeny of Armillaria based on a combined ITS, IGS and tef1 sequence data (Fig 2, Table 2). However, insufficient data are available for the LSU gene region in GenBank. Then, it is difficult to have comparative phylogenetic analyses but the single gene analysis of each gene was carried out to compare the topology of the tree and clade stability. This phylogenetic tree is largely in accordance with earlier studies from Coetzee et al. (2018) and provides the most conclusive phylogeny of the genera to date. Genealogical concordance phylogenetic species recognition (GCPSR) using the concordance among several gene trees (Taylor et al. 2000; Dettman et al. 2003) to delineate species has become standard in fungal taxonomy. However, except for a few studies (Guo et al. 2016; Tsykun et al. 2013), this taxonomic method has not been widely implemented in Armillaria taxonomy. Sequences of the genomes of key species are already providing prospects to study the evolution and systematics of Armillaria. They are certain to lead to important breakthroughs regarding not only the taxonomy but the biology and ecology of these fungi in the future (Sipos et al. 2017).
Recommended genetic marker (genus level)– ITS
Recommended genetic markers (species level)– ITS, IGS1, tef1
Additional genetic markers (species level)– LSU, tub2
Accepted number of species– There are 278 epithets in Index Fungorum (2020) listed for this genus. However, sequence data are only available for 31 species including 16 groups of unnamed species (Table 1).
References– Watling et al. 1991, Pegler 2000, Harrington and Wingfield 1995 (morphology); Coetzee et al. 2000a,b, 2001a,b, Maphosa et al. 2006, Mwenje et al. 2006, Kim et al. 2006, Coetzee et al. 2018 (molecular phylogeny).
Table 1 DNA barcodes available for Armillaria. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Sources | Country | ITS | LSU | tef1 | IGS1 |
| Armillaria affinis | JMCR.126 | Central America? | – | AF261356 | – | – |
| A. altimontana | POR100* | USA | AY213579, AY213580 | – | JN944606, JF313117 | AY509181 |
| A. aotearoa | NZFRI-M 5283* | New Zealand | NR_151846 | – | KU295542 | – |
| A. borealis | CMW31075 | Belarus | – | – | KM205252 | KM205305 |
| CMW31072 | China | – | – | KM205251 | KM205304 | |
| HKAS 76263, Gt571 | China | KT822294 | – | KT822426 | – | |
| BRNM 699842, MUAF 501 | Czech Republic | EU257713 | – | EU251402 | EU257708 | |
| CMW3172 | Finland | – | DQ338540 | DQ435623 | – | |
| A1 | Finland | JN657467 | – | JN657494 | JN657440 | |
| A5 | Germany | JN657468 | – | JN657495 | JN657441 | |
| A618 | Switzerland | JN657469 | – | JN657496 | JN657442 | |
| A. calvescens | ST3 | USA | AY213559 | JF895899 | JF313138, JF895835 | AY509163 |
| ST17 | USA | AY213560, AY213561 | JF895900 | JF313130, JF895836 | AY509164 | |
| TH DJA 91/PUL F2895 | Cameroon | KU170952 | KU170942 | KU289112 | – | |
| A. cepistipes | BRNM 706814, MUAF 516 | Czech Republic | EU257715 | – | EU251395 | EU257709 |
| M110 | Canada | AY213581 | – | JF313121 | AY509182 | |
| HKAS 86583, 01108/1 | China | KT822290 | – | KT822417 | – | |
| B3 | Finland | JN657445 | – | JN657472 | JN657418 | |
| B5 | Italy | JN657446 | – | JN657473 KJ414321 | JN657419, KJ414318 | |
| 94-39-04 | Japan | AB510853 | – | AB510786 | AB510809 | |
| SY1Ra | UK? | – | – | JF746917 | JF288720 | |
| C5C-S1 | Ukraine | JN657450 | – | JN657477 | JN657423 | |
| W113 | USA | AY213583 | – | JF313115 | AY509184 | |
| BRNM 695717 | Czech Republic | EU257716 | – | EU251396 | EU257710 | |
| HKAS 86586, 97033/1 | China | KT822263 | – | KT822416 | – | |
| KFRI1616 | Korea | MG543860 | – | MG544785 | – | |
| A. ectypa | BRNM 704974 | Austria | EU257720 | – | EU251403 | EU257712 |
| CMW15693 | France | – | DQ338547 | FJ875698 | – | |
| HKAS 86565, 70011/13 | France | KT822340 | – | KT822438 | – | |
| TFM27105, Je-2 | Japan | AB559000 | – | AB558992 | – | |
| A. fumosa | CMW4957, 123 | Australia | AF329917 | DQ338552 | DQ435646 | – |
| A. fuscipes | CBS 118122, CMW5844, WG1I | Ethiopia | AY882969 | – | – | AY172032 |
| CMW7184 | Kenya | AY882973 | – | – | AY882965 | |
| CMW4953 | La Reunion | AY882974 | DQ338556 | DQ435622 | AY882963 | |
| CMW4871 | Malawi | AY882976 | – | – | AY882959 | |
| CMW2717 | South Africa | AY882971 | – | – | AF204821 | |
| CMW4949 | Tanzania | AY882978 | – | – | AY882961 | |
| CMW4874 | Zimbabwe | AY882967 | – | – | AF489481 | |
| A. gallica | M70 | Canada | AY213568 | – | JF313123 | AY509171 |
| CMW31087 | China | – | – | KM205260 | KM205313 | |
| HKAS 86569, 93421/1 | China | KT822277 | – | KT822414 | – | |
| BRNM 706835, MUAF 575 | Czech Republic | EU257718 | – | EU251390 | EU636240 | |
| E4 | France | JN657452 | – | JN657479 | JN657425 | |
| 86-016/3 | Germany | – | – | KJ200952 | KJ200946 | |
| 86-008/2 | Iran | – | – | KJ200954 | KJ200948 | |
| NA4 | Japan | AB510881 | – | AB510761 | AB510834 | |
| MEX55 | Mexico | JX281809 | – | KC111014 | JX281799 | |
| CMW7202 | South Africa | AY190247 | – | – | AY190245 | |
| HY2a | Ukraine | JN657455 | – | JN657482 | JN657428 | |
| Aga235 | USA | – | JF895911 | JF895847 | – | |
| ST22, EL-1 | USA | AY213569, AY213570 | JF895912 | JF313126, JF895848 | AY509172 | |
| Ame10 | Korea | MG543850 | – | MG544774 | – | |
| KA14-1647 | Korea | MG543859 | – | MG544784 | – | |
| Ame7 | Korea | MG543852 | – | MG544777 | – | |
| A. gemina | JB-38A | Canada | FJ664586 | FJ618757 | FJ618670 | – |
| ST8 | USA | AY213555 | – | JF313136 | AY509158, AY509159 | |
| A. heimii | C4 | Congo | AY333917 | – | – | AY330630 |
| 166 | – | AY333913 | – | – | AY330634 | |
| K59 | Kenya | AY333916 | – | – | AY330627 | |
| A. hinnulea | CMW4980 | Australia | – | DQ338555 | DQ435648 | AF445077 |
| CMW4990 | New Zealand | AF329905 | DQ338555 | DQ435648 | – | |
| A. jezoensis | HUA9116 | Japan | – | – | – | D89921 |
| A. limonea | CMW4680 | New Zealand | AF329930 | DQ338560 | DQ435655 | AF445073 |
| A. lutea | 90-4 (Alut) | USA | – | – | – | AF243066 |
| A. luteobubalina | CMW4977 | Australia | AF329912 | DQ338559 | DQ435657 | AF445069 |
| A. mellea | AFTOL-ID449 | USA | AY789081 | AY700194 | AY881023 | – |
| B176 | England | AF163578 | – | – | AF163602 | |
| HKAS 86590, 00020/6 | China | KT822251 | – | KT822354 | – | |
| D1 | France | JN657464 | – | JN657491 | JN657437 | |
| B1212, CMW4615, 94056/1 | Hungary | AF163581 | – | – | AF163605 | |
| B1205, CMW4613, 86009/1 | Iran | AF163583 | – | DQ435637 | AF163606 | |
| CBS122232, CMW11265, 426 | Italy | FJ875692 | FJ875694 | DQ435636 | – | |
| FFPRI420861, WD2588, 89-07 | Japan | AB510852 | – | AB510796 | AB510808 | |
| HKAS 86598, PFD84-103 | Kenya | KT822248 | – | KT822348 | – | |
| MEX74 | Mexico | JX281807 | – | KC111011 | JX281797 | |
| CMW3975 | South Africa | AF310329 | – | – | AF310327 | |
| B916, CMW4610, A-5 | South Korea | AF163592 | – | DQ435639 | AF163612 | |
| HY-3 | Ukraine | JN657466 | – | JN657493 | JN657439 | |
| Am115 | USA | – | JF895920 | JF895856 | – | |
| B927 | USA | AF163595 | FJ875695 | DQ435634 | AF163608 | |
| CMW31161 | China | – | – | KM205267 | KM205320 | |
| CMW8082 | Bhutan | AY554333 | – | – | AY554335 | |
| HUA93110 | Japan | – | – | – | D89922 | |
| A. mexicana | MEX87* | Mexico | KR061310 | – | KR061314 | KR061306 |
| A. montagnei (ex luteobublina) | CMW5446 | Argentina | AF448422 | DQ338562 | DQ435650 | AF445068 |
| A. montagnei (ex luteobublina) | CMW8876 | Chile | AF448423 | – | DQ435658 | AF445065 |
| A. montagnei (Lineage II) | Arg309 | Argentina | FJ660939 | FJ711625 | – | – |
| A. montagnei (Lineage II) | Arg270* | Argentina | FJ711609 | – | – | – |
| A. nabsnona | ST16 | USA | AY213574 | – | JF313124 | AY509178 |
| HKAS 85523, Gt798 | China | KT822333 | – | KT822411 | – | |
| M90 | Canada | AY213573 | – | JF313122 | AY509176, AY509177 | |
| 00-3-1 | Japan | AB510899 | – | AB510766 | AB510850 | |
| C21 | USA | AY213572 | – | JF313119 | AY509174, AY509175 | |
| CMW6905 | USA | – | DQ338542 | DQ435631 | – | |
| Juk14411 | Korea | MG543857 | – | MG544782 | – | |
| A. novae-zelandiae | CMW4967 | Australia | AF329921 | – | DQ435651 | – |
| CMW5448 | Chile | AF448417 | DQ338554 | DQ435653 | – | |
| CMW4143 | Indonesia | AF448421 | DQ338564 | DQ435654 | – | |
| CMW3951 | Malaysia | AF448419 | DQ338553 | – | – | |
| CMW4722 | New Zealand | AF329926 | DQ338551 | DQ435652 | – | |
| A. novae-zelandiae (Lineage I) | Arg49 | Argentina | FJ660935 | FJ711629 | – | – |
| A. ostoyae | SP308014* | Brazil | EF639348 | – | – | – |
| BRNM706815 | Czech Republic | EU257717 | – | EU251400 | EU257711 | |
| CMW31102 | China | – | – | KM205272 | KM205325 | |
| HKAS 86579, 96043/11 | China | KT822310 | – | KT822428 | – | |
| C2 | France | JN657459 | – | JN657486 | JN657432 | |
| 88-01-19 | Japan | AB510859 | – | AB510784 | AB510815 | |
| D20 | Switzerland | JN657463 | – | JN657490 | JN657436 | |
| HpAg1 | Ukraine | JN657462 | – | JN657489 | JN657435 | |
| P1404 | USA | AY213554 | – | JF313140 | AY509157 | |
| Ame5 | Korea | MG543851 | – | MG544776 | – | |
| 3626 | Australia | FJ664607 | FJ618752 | FJ618665 | – | |
| A. puiggarii | MCA 3111/PUL F2896/BRG 41295 | Guyana | KU170954 | KU254228 | KU289104 | – |
| A. sinapina | V48.5 | Canada | FJ664609 | FJ618763 | FJ618676 | – |
| M50 | Canada | AY213563, AY213564 | – | JF313114 | AY509167 | |
| CMW31112 | China | – | – | KM205277 | KM205330 | |
| HKAS 86566, 96015/39 | China | KT822323 | – | KT822422 | – | |
| 96-7-1 | Japan | AB510873 | – | AB510774 | AB510827 | |
| P2-7 | USA | – | JF895916 | JF895850 | – | |
| ST12 | USA | AY213565 | – | JF313132 | AY509168 | |
| A. singula | HUA9101* | Japan | – | – | – | D89926 |
| A. socialis | T2 | France | DQ784801 | – | – | – |
| A. solidipes | MS2-11 | USA | – | JF895918 | JF895852 | – |
| CMW31107 | Finland | – | – | KM205275 | KM205328 | |
| A. sparrei | PSpa86.5 | Argentina | FJ664612 | FJ618750 | – | – |
| Arg12 | Argentina | FJ660948 | FJ711618 | – | – | |
| A. umbrinobrunnea | Arg25 | Argentina | FJ660946 | FJ711621 | – | – |
| A. tabescens | CMW31118 | China | – | – | KM205280 | KM205333 |
| 99122/13 | China | KT822339 | – | KT822441 | – | |
| CMW3165 | France | – | DQ338546 | DQ435642 | – | |
| CMW31119 | Italy | – | – | KM205281 | KM205334 | |
| HKAS 86604, CT1097.3 | Italy | KT822338 | – | KT822440 | – | |
| 96-1-8 | Japan | AB510867 | – | AB510804 | AB510823 | |
| HKAS 86605, 901582 | Slovenia | KT822337 | – | KT822439 | – | |
| HAt1S5 | Ukraine | HQ232292 | – | HQ285906 | HQ232284 | |
| ATMUS2 | USA | AY213588 | – | JF313113 | AY509189, AY509190 | |
| Bhutanese Group 2 | CMW10581 | Bhutan | AY554329 | – | FJ875699 | AY624365 |
| Chinese Biological species C | CMW31123 | China | – | – | KM205284 | KM205337 |
| CMW31124 | China | – | – | KM205285 | KM205337 | |
| Chinese Biological species F | CMW31127 | China | – | – | KM205286 | KM205339 |
|
|
CMW31128 | China | – | – | KM205287 | KM205340 |
| Chinese Biological species H | CMW31136 | China | – | – | KM205293 | KM205346 |
| CMW31138 | China | – | – | KM205294 | KM205347 | |
| Chinese Biological species J | CMW31140 | China | – | – | KM205296 | KM205349 |
| CMW31142 | China | – | – | KM205297 | KM205350 | |
| Chinese Biological species L | CMW31144 | China | – | – | KM205298 | KM205351 |
| CMW31145 | China | – | – | KM205299 | KM205352 | |
| Chinese Biological species N | CMW31146 | China | – | – | KM205300 | KM205353 |
| CMW31148 | China | – | – | KM205301 | KM205354 | |
| Chinese Biological species O | CMW31150 | China | – | – | KM205302 | KM205355 |
| CMW31151 | China | – | – | KM205303 | KM205356 | |
| Chinese Lineage 1 | HKAS 86615 | China | KT822315 | – | KT822384 | – |
| HKAS 86621 | China | KT822306 | – | KT822386 | – | |
| Chinese Lineage 2 | HKAS 86623 | China | KT822318 | – | KT822363 | – |
| HKAS 86551 | China | KT822279 | – | KT822367 | – | |
| Chinese Lineage 3 | HKAS 86613 | China | KT822319 | – | KT822388 | – |
| HKAS 86614 | China | KT822305 | – | KT822391 | – | |
| Chinese Lineage 4 | HKAS 86602 | China | KT822308 | – | KT822378 | – |
| HKAS 86606 | China | KT822281 | – | KT822359 | – | |
| Chinese Lineage 5 | HKAS 86574 | China | KT822324 | – | KT822361 | – |
| HKAS 86574 | China | KT822327 | – | KT822364 | – | |
| Chinese Lineage 6 | HKAS 86570 | China | KT822320 | – | KT822402 | – |
| HKAS 86571 | China | KT822288 | – | KT822404 | – | |
| Chinese Lineage 7 | HKAS 83303 | China | KU378047 | – | KT822437 | – |
| HKAS 83361 | China | KU378048 | – | KT822436 | – | |
| Japanese Nag. E | 94-2-1 | Japan | AB510888 | – | AB510768 | AB510840 |
| 2000-71-13 | Japan | AB510879 | – | AB510773 | AB510832 |
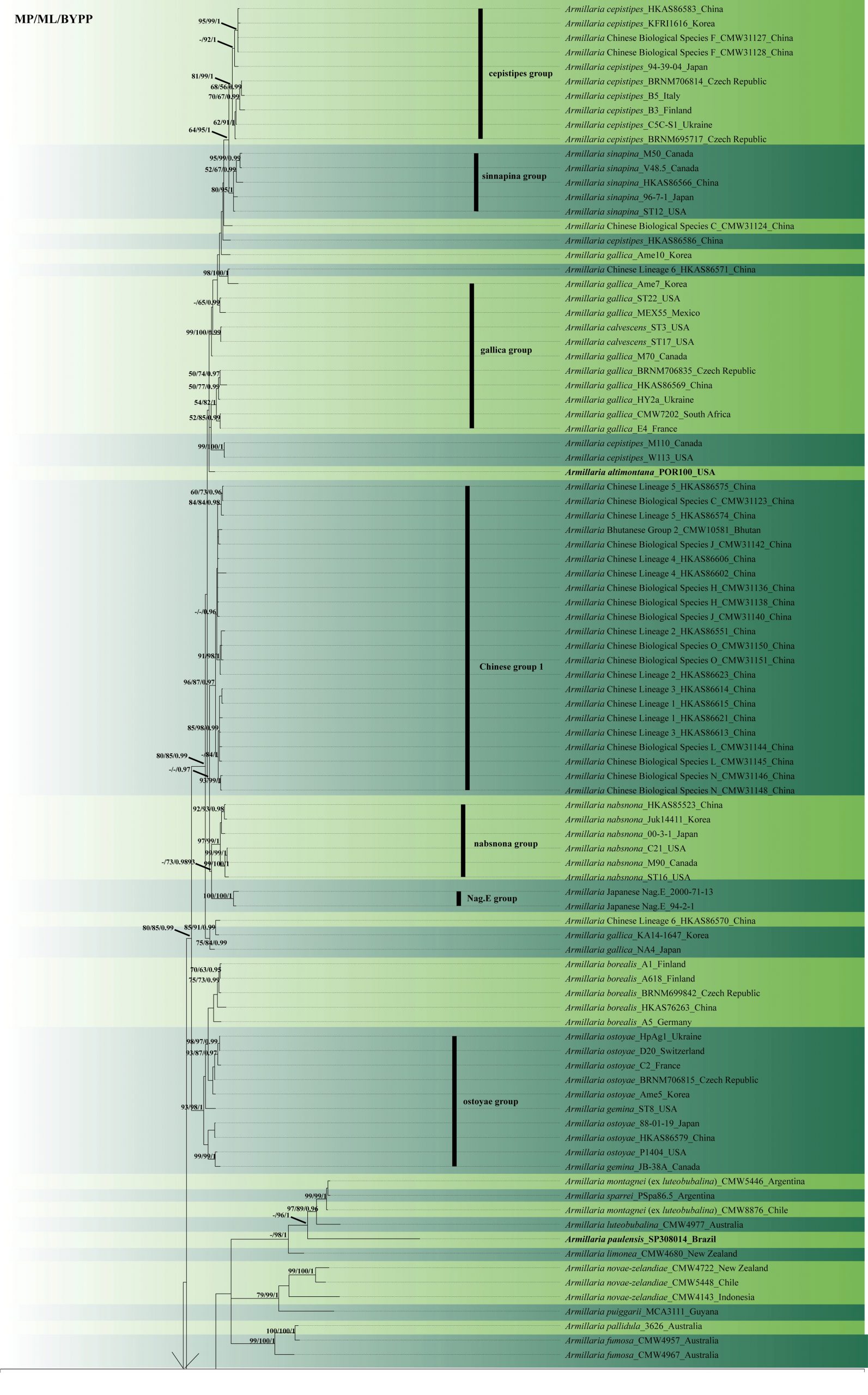
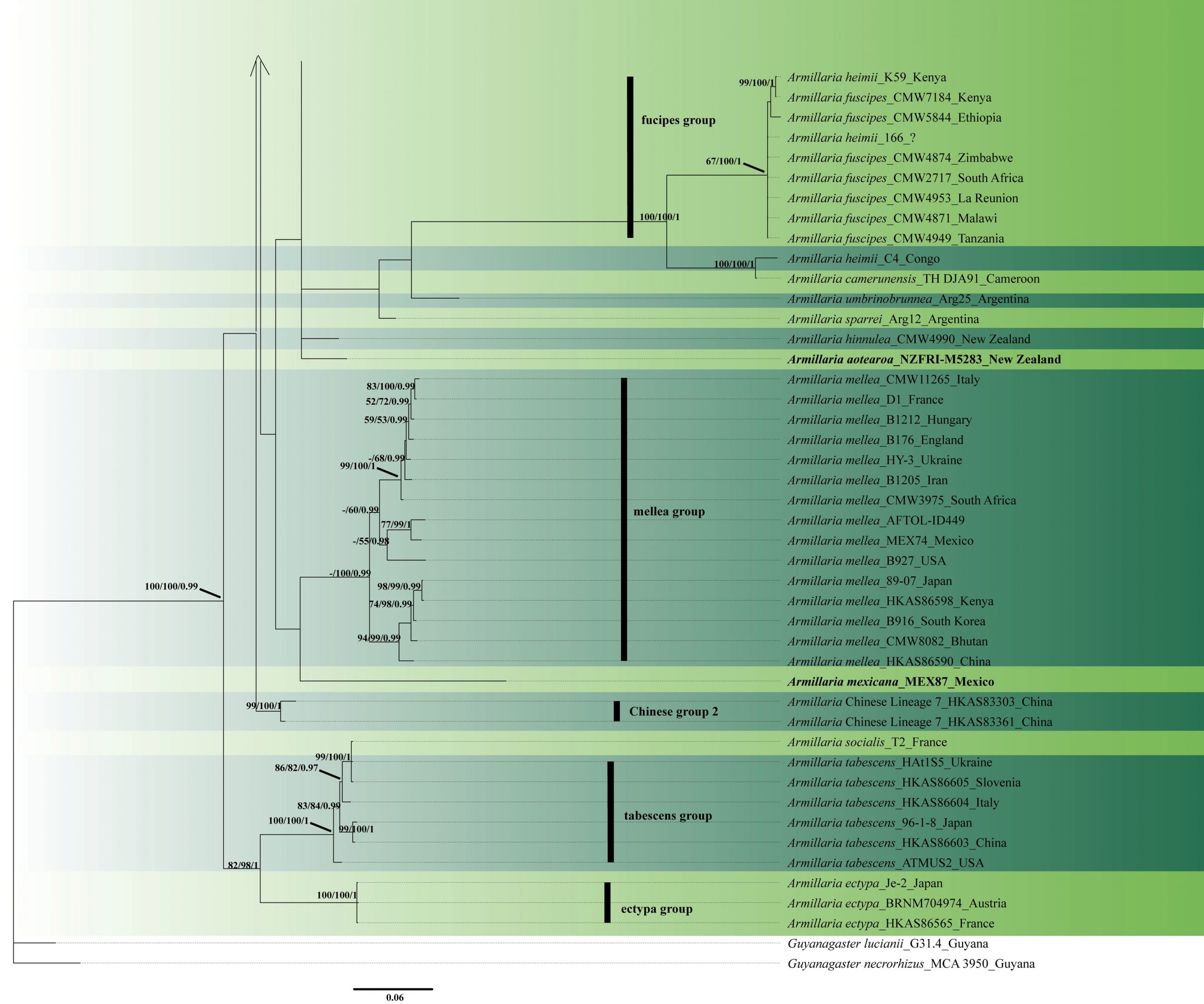
Fig. 2 Phylogenetic tree generated by maximum likelihood analysis of combined ITS-IGS-tef1 sequence data of Armillaria species. Related sequences were obtained from GenBank. One hundred and thirty-nine strains are included in the analyses, which comprise 4557 characters including gaps. The tree was rooted with Guyanagaster lucianii (G31.4) and Guyanagaster necrorhizus (MCA 3950). Single gene analyses were carried out to compare the topology of the tree and clade stability. Tree topology of the ML analysis was similar to the MP and BYPP. ML phylogenetic tree inference was performed using RAxML version 8.2.12 on the CIPRES web server, using a mixed-model analysis and the GTRCAT model of substitution. The four partitions were defined as ITS, IGS, tef1 exons and tef1 introns. The best scoring RAxML tree with a final likelihood value of −25308.198187 is presented. The matrix had 1957 distinct alignment patterns, with 65.74% of undetermined characters or gaps. Estimated base frequencies of ITS were as follows: A =0.227071, C =0.203923, G =0.235701, T =0.333305; substitution rates AC =0.628852, AG=3.751709, AT =1.365607, CG =1.467905, CT =2.788595, GT = 1.000000. Estimated base frequencies of IGS were as follows: A =0.244624, C =0.196588, G =0.242370, T =0.316418; substitution rates AC =0.954911, AG=3.055115, AT =1.041498, CG =1.278095, CT = 3.421100, GT = 1.000000. Estimated base frequencies of tef1 exons were as follows: A =0.228587, C =0.301128, G =0.255865, T =0.214420; substitution rates AC =0.905728, AG=3.660986, AT =1.564184, CG =0.648739, CT = 28.048363, GT = 1.000000. Estimated base frequencies of tef1 introns were as follows: A =0.215042, C =0.222693, G =0.185633, T =0.376631; substitution rates AC =1.170263, AG=5.878084, AT =0.847943, CG =1.087990, CT = 5.095797, GT = 1.000000; gamma distribution shape parameter α =0.1000000000. The maximum parsimonious dataset consisted of 2908 constant, 1172 parsimony-informative and 477parsimony-uninformative characters. The parsimony analysis: CI = 0.610, RI = 0.861, RC = 0.525, HI = 0.390 in the first tree. Bayesian posterior probability was performed using the Markov chain Monte Carlo (MCMC) method implemented in MrBayes 3.2.6 with a mixed-model partition identical to the ones defined in the ML analysis. The best-fit nucleotide substitution model was separately determined for each partition with jModeltest version 2.1.10 on CIPRES, using the Akaike Information Criterion. K80+I, K80+I, SYM+G and HKY+G were selected as best-fit models for ITS, IGS, tef1 exons and tef1 introns, respectively. At the end of the runs, the average deviation of split frequencies was 0.016675. MP and RAxML bootstrap support value ≥ 50% and BYPP ≥0.95are shown, respectively, near the nodes. Holotype or ex-type strains are in bold.

No Comments