24 Oct Clinoconidium
Clinoconidium Pat., Bulletin de la Société Mycologique de France 14: 156 (1898)
Background
Clinoconidium is an important genus that causes smut disease on plants in the family Lauraceae. This genus was established by Patouillard (1898) and typified with Clinoconidium farinosum. Taxonomically, Clinoconidium is placed in Cryptobasidiaceae (Exobasidiales, Exobasidiomycetes, Basidiomycota) and characterized by aseptate, colourless, and globose to ovoid basidiospores which are dispersed individually. The name Clinoconidium was considered illegitimate because of the designation of an illegitimate type species name; however, it was later validated by Saccardo (1902).
Clinoconidium is a gall producing genus which was once named as Ustilago by Ito (1935, 1936) due to the presence of a powdery spore mass on the surface of the galls. This genus was also transferred to another gall producing genus Melanopsichium by Kakishima (1982). However, it was renamed as Clinoconidium as its sorus structure and spore features are quite different from those of Ustilago (Saccardo 1902). The spores of Ustilago species are formed from sporogenous hyphae, whereas this fungus produces spores from hymenial layers in the galls. Spore walls are comparatively thinner than those of Ustilago. The differentiation from Melanopsichium, a gall producing taxon on plants in Polygonaceae (Vánky 2013) includes variation in gall structures and sporulation. Melanopsichium produces spores in chambers formed inside of gall tissues, while this species produces spores in peripheral lacunae on the surface of gall tissues. The morphological characters of these taxa showed its close similarity to Clinoconidium.
Classification – Basidiomycota, Ustilaginomycotina, Exobasidiomycetes, Exobasidiomycetidae, Exobasidiales, Cryptobasidiaceae
Type species – Clinoconidium farinosum Pat. ex Sacc. & P. Syd
Distribution – Brazil, China, Costa Rica, India, Japan, Panama, Spain, Taiwan and Venezuela
Disease symptoms – mainly observed as powdery pappus gall in fruits. Infection initiates on very young fruits, converted into round, wrinkled galls. The fruit galls are then covered with a powdery mass of spores during early days of infection, withering in the rainy season, leaving behind hard, earthy, brown galls. On Cinnamon, entire young fruits are molded with buff and spongy smut like taxa in the full bloom of disease. Interestingly this infection is restricted to fruits only (Fig. 1).
Hosts – different plants of Lauraceae including, Apollonias barbujana, Cinnamomum burmannii, C. camphora, C. daphnoides, C. tamala, C. tenuifolium, Nectandra sp., Octea sp., Oreodaphne sp. and Phoebe neurophylla (Farr and Rossman 2020).
Fig. 1 Clinoconidium sp. on Cinnamomum sp. a host plant with infected and healthy fruits, b healthy fruits, c–d infected fruits at various stages of infection
Morphological based identification and diversity
This is an important pathogenic genus; producing galls on shoot buds of host plants belonging to the family Lauraceae. Fruits of the host are completely or partially transformed into reddish-brown to dark brown, irregularly malformed, enlarged, globose to subglobose galls; larger than normal fruits. Hymenia formed in peripheral lacunae of the galls are pale yellow to whitish and covered by the host epidermis. Inner tissues of galls consist of hyphae and deformed plant cells. Hyphae are intercellular, hyaline, compact, septate, smooth-walled and lack clamp connections, while haustoria are intercellular, slightly lobed to irregular and observed in deformed host cells. Upon maturation, galls rupture, exposing orange to dark brown or creamish white spore masses which cover the entire infected young fruits. Sterile hyphae can be found intermingled between the basidia in some species and are indistinguishable from young basidia or absent in some species of Clinoconidium. Basidia are clavate, hyaline, depressed, difficult to observe and gastroid, densely aggregated in masses, formed in irregular fascicles from basally agglutinated hyphae and the wall is densely foveolate when mature. Basidiospores are ellipsoid, clavate, pyriform, fusoid, globose, subglobose to oval, aggregated in a creamish white to brown coloured masses on the surface of the galls, hyaline or wall pale brown to brown, rugose when mature; producing long branched hyphae with septa when germinated on culture media and proliferating sympodially.
Molecular based identification and diversity
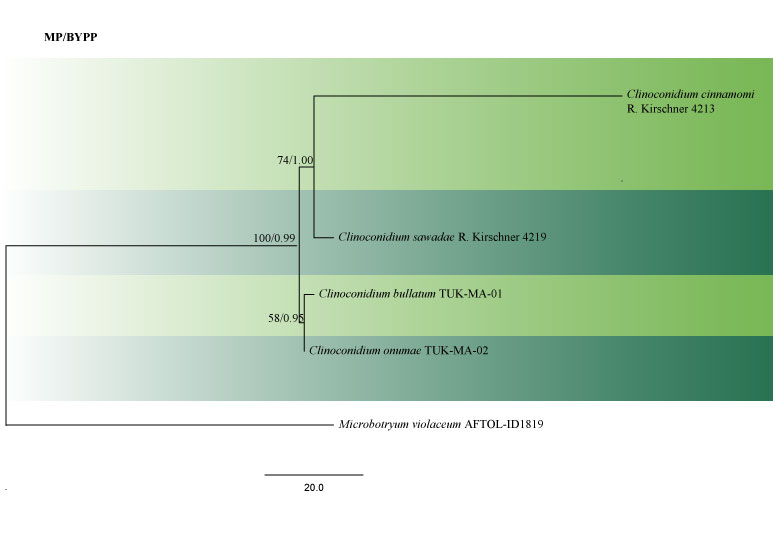
There are seven epithets of Clinoconidium recorded on various plant hosts. Sequence data for Clinoconidium bullatum, C. cinnamomi, C. onumae and C. sawadae are available in GenBank, including sequence data for LSU and ITS. Clinoconidium farinosum and C. globosum lack sequence data in GenBank. ITS and LSU are the most suitable loci for delineation of species within the genus (Fig 2).
Recommended genetic markers (genus level) – ITS, LSU
Recommended genetic markers (species level) – ITS, LSU
Accepted number of species – There are seven species epithets in Index Fungorum (2020), however, only four species have DNA molecular data (Table 1).
References – Hendrichs et al. 2003, Jiang and Kirschner 2016, Kakishima et al. 2017a,b (morphology, phylogeny)
Table 1 DNA barcodes available for Clinoconidium.
| Species name | Strain Name | ITS | LSU |
| Clinoconidium bullatum | TUK-MA-01 | – | AB178259 |
| C. cinnamomi | R. Kirschner 4213 | KX196602 | KX196604 |
| C. onumae | TUK-MA-02 | – | AB178260 |
| C. sawadae | R. Kirschner 4219 | KX196600 | KX196603 |
Fig. 2 Phylogram generated from MP analysis based on combined sequences of LSU and ITS sequences of all the species of Clinoconidium with molecular data. Related sequences were obtained from GenBank. Five taxa are included in the analyses, which comprise 1100 characters including gaps, of which 910 characters are constant, 182 characters are parsimony-uninformative, eight characters parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 202 steps (CI = 0.980, RI 0.500, RC = 0.490, HI = 0.020) in the first tree Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Microbotryum violaceum (AFTOL-ID1819). Maximum parsimony bootstrap support value ≥50% and BYPP ≥0.9 are shown respectively near the nodes.


No Comments