15 Oct Verticillium
Verticillium Nees, Syst. Pilze (Würzburg): 57 (1816) [1816-17]
The genus Verticillium Nees was introduced by Nees von Esenbeck (1816) for a single saprotrophic species, V. tenerum Nees, which was proposed as the type species. Zare et al. (2004) synonymized V. tenerum under Acrostalagmus luteoalbus (Link) Zare, W. Gams & Schroers, and Gams et al. (2005) proposed V. dahliae Kleb. as the conserved type of the genus Verticillium.
Verticillium includes several plant pathogenic species that infect trees, insects, mushrooms and, in particular, dicotyledonous plants: V. dahliae Kleb., V. albo-atrum Reinke et Berth., V. nigrescens Pethybr., V. nubilum Pethybr., V. tricorpus Isaac., V. theobromae (Turc.) Mas. & Hughes and V. fungicola (Preuss) Hassebrauk (Pegg and Brady 2002). Zare et al. (2007) assigned V. nigrescens to the genus Gibellulopsis and V. theobromae to Musicillium. Zare and Gams (2008) assigned V. fungicola to the genus Lecanillium. Verticillium dahliae and V. albo-atrum are the two most notorious species which cause Verticillium wilt diseases in a wide range of mainly dicotyledonous hosts and result in billions of dollars of damage annually in crop losses worldwide (Pegg and Brady 2002; Barbara and Clewes 2003; Inderbitzin et al. 2011).
Verticillium species, in particular, V. dahliae and V. albo-atrum, can infect a wide range of plant species. Many hosts of Verticillium species were given by different mycologists, e.g., Van der Meer (1925); Rudolph (1931); Engelhard and Carter (1956); Parker (1959); Stark (1961); Devaux and Sackston (1966); Himelick (1969), including high-value crop plants, e.g., cotton (Land et al. 2016), lettuce (Garibaldi et al. 2007; Powell et al. 2013), mango (Baeza-Montanez et al. 2010; Ahmed et al. 2014), gold kiwifruit (Auger et al. 2009), bean (Berbegal and Armengol 2009; Sun et al. 2016; Blomquist et al. 2017), watermelon (Bruton et al. 2007), olive tree (Lo Giudice et al. 2010; Kaliterna et al. 2016), potato (Pace-Lupi et al. 2006), pumpkin (Rampersad 2008). The most comprehensive hosts’ list was provided by Pegg and Brady (2002).
Classification – Sordariomycetes, Hypocreomycetidae, Glomerellales, Plectosphaerellaceae
Type species – Verticillium dahliae Kleb., Mykol. Zentbl. 3: 66 (1913)
Distribution – Worldwide
Disease symptoms – Wilt
Wilt caused by Verticillium species is a serious fungal disease that causes injury or death to many plant species. Symptoms of this disease vary according to the host species and to environmental conditions including sudden wilting of small branches, yellowing of foliage, stunt growth and premature defoliation. Olive- green to black streaks can be observed in the sapwood of infected branches. In cross-sections, vascular tissue appears as a dark ring or pinpoint dark spots. Initial symptoms can occur on one side of the tree or the entire plant (Fradin and Thomma 2006; Blum et al. 2018).
Hosts – Species of this genus have a broad host range including members of Amaranthaceae, Amaryllidaceae, Asteraceae, Brassicaceae, Musaceae, Pinaceae, Rosaceae, Rubiaceae, Sapindaceae, Solanaceae, Theaceae, Vitaceae and Zingiberaceae (Farr and Rossman 2019).
Morphological based identification and diversity
There are 270 records in Index Fungorum (2019) and 288 records in MycoBank (Crous et al. 2004) in this genus. Most of the species have been transferred to other genera, e.g., Gibellulopsis (Zare et al. 2007), Haptocillium (Zare and Gams 2001b), Lecanicillium (Zare and Gams 2001a), Musicillium (Zare et al. 2007), Pochonia (Zare et al. 2001a), Simplicillium (Zare and Gams 2001a). Only ten species, V. albo-atrum, V. alfalfae, V. dahliae, V. isaacii, V. klebahnii, V. longisporum, V. nonalfalfae, V. nubilum, V. tricorpus and V. zaregamsianum, were accepted within Verticillium sensu stricto (Inderbitzin et al. 2011). No sexual states are known. Descriptions and a key to these ten species were provided by Inderbitzin et al. (2011).
Molecular based identification and diversity
The first phylogenetic analysis of Verticillium species was made by Morton et al. (1995) to analysis the relationship between V. alboatrum and V. dahliae based on the internal transcribed spacer regions and intervening 5.8S rDNA (ITS) sequences data. Subsequently, SSU, LSU, RNA polymerase II largest subunit gene (RPB1), the cytochrome oxidase subunit III gene (cox3), the small ribosomal rRNA subunit (rns), NADH dehydrogenase subunit genes (nad1 and nad3) were used (Barbara and Clewes 2003; Pantou et al. 2005; Zare and Gams 2008 2016).
Recommended genetic markers (genus level) – LSU
Recommended genetic markers (species level) –ITS
Accepted number of species: 10 species.
References: Inderbitzin et al. 2011 (morphology and phylogeny); Barbara and Clewes 2003, Morton et al. 1995, Pantou et al. 2005, Zare and Gams 2008, 2016 (phylogeny); Fradin and Thomma 2006, Blum et al. 2018 (pathogenicity)
Table GenBank accession numbers of Verticillium isolates included in this study
| Taxa | Strains | haplotype | ACT | tef1 | GPD | TS |
| Verticillium albo-atrum | PD747 | JN188144 | JN188272 | JN188208 | JN188080 | |
| V. alfalfa | PD489 | JN188097 | JN188225 | JN188161 | JN188033 | |
| V. dahlia | PD322 | HQ206921 | HQ414624 | HQ414719 | HQ414909 | |
| V. isaacii | PD660 | HQ206985 | HQ414688 | HQ414783 | HQ414973 | |
| V. klebahnii | PD401 | JN188093 | JN188221 | JN188157 | JN188029 | |
| V. longisporum | PD348 | a1 | HQ206930 | HQ414633 | HQ414728 | HQ414918 |
| V. longisporum | PD348 | d1 | HQ206931 | HQ414634 | HQ414729 | HQ414919 |
| V. longisporum | PD356 | a1 | HQ206934 | HQ414637 | HQ414732 | HQ414922 |
| V. longisporum | PD356 | d2 | HQ206935 | HQ414638 | HQ414733 | HQ414923 |
| V. longisporum | PD687 | a1 | HQ206993 | HQ414696 | HQ414791 | HQ414981 |
| V. longisporum | PD687 | d2 | HQ206994 | HQ414697 | HQ414792 | HQ414982 |
| V. nonalfalfae | PD592 | JN188099 | JN188227 | JN188163 | JN188035 | |
| V. nubilum | PD742 | JN188139 | JN188267 | JN188203 | JN188075 | |
| V. tricorpus | PD690 | JN188121 | JN188249 | JN188185 | JN188057 | |
| V. zaregamsianum | PD736 | JN188133 | JN188261 | JN188197 | JN188069 |
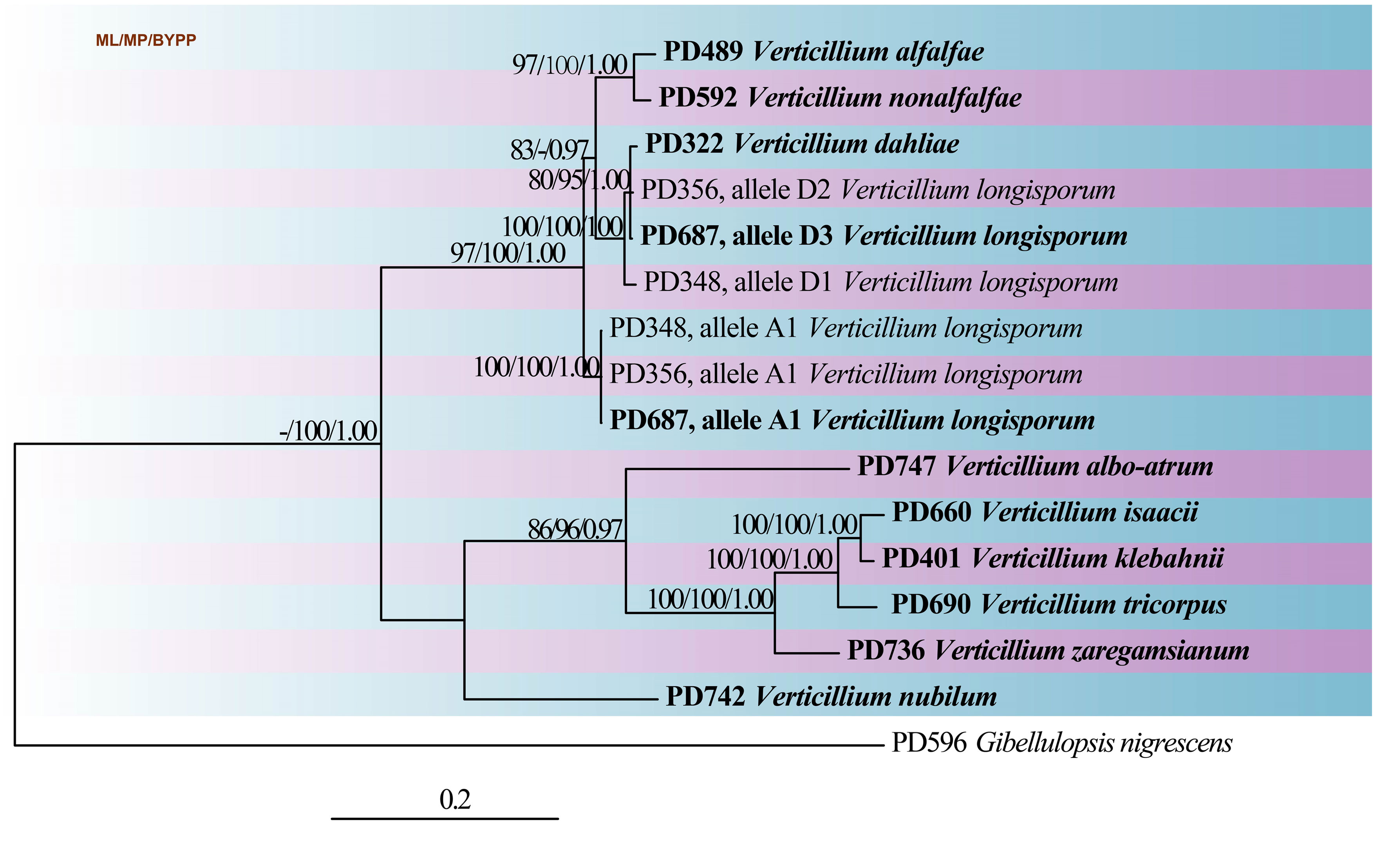
Fig Phylogenetic tree generated from Bayesian analysis based on combined ACT, tef1, GPD and TS sequence data for the genus Verticillium. Sixteen strains are included in the combined genes sequence analyses which comprise total of 2597 characters (609 characters for the ACT, 620 characters for tef1, 744 characters for GPD, 624 characters for TS) after alignment. For the Bayesian analysis, two parallel runs with six chains were run for 1,000,000 generations and trees were sampled every 100th generation, resulted in 20002 trees from two runs of which 15002 trees were used to calculate the posterior probabilities (each run resulted in 10001 trees of which 7501 trees were sampled). Bootstrap support values for maximum likelihood (ML, first set) and maximum parsimony (MP, second set) greater than 75% and Bayesian posterior probabilities greater than 0.95 are indicated above or below the nodes. Ex-type strains are in bold. The tree is rooted with Gibellulopsis nigrescens (PD596).

No Comments