26 Oct Ganoderma
Ganoderma P. Karst., Revue mycol., Toulouse 3(no. 9): 17 (1881)
Background
Ganoderma was established by Karsten (1881) based on G. lucidum and characterized by double-walled basidiospores with truncate apices and ornamented endospores, and a crusty or shiny pileus surface (Moncalvo and Ryvarden 1997). This genus was divided into two subgenera, Ganoderma and Elfvingia by Karsten (1889). Various authors used different taxonomic characters for the identification of species (e.g., Murrill 1902, 1903; Atkinson 1908; Coleman 1927; Corner 1947), which resulted in an intricate taxonomy, with 344 species names in speciesfungorum.org, but an estimated 180 species (He et al. 2019) and Steyaert (1972, 1980) worked extensively on the genus and introduced many new species, transferred many to the genus and removed several synonyms. Ryvarden (1985) and Gottlieb and Wright (1999a,b) studied the macro- and micromorphology. Ganoderma presently comprises sections Amauroderma and Ganoderma, subgenera: Ganoderma and Trachyderma (Index Fungorum 2020, Wijayawardene et al. 2020).
Relevant characteristics for Ganoderma species delimitation are based on the macro and micromorphological characteristics (see in Fig. 1). The basidiomes are annual or perennial, dimidiate, sessile or substipitate to stipitate, with distinctive non-laccate (dull) or weakly to strongly laccate, glossy, shiny, smooth, spathulate, furrows, which are sulcate on the pileus surface. Some strains have several layers of thick, dull cuticles or shiny, with thin cuticle or cuticle of clavate end cells. The context is cream to dark purplish brown, brown to dark brown, sometimes spongy to firm-fibrous. Pores are 4–7 per mm, angular, entire, subcircular to circular, regular, mostly cream or white when young, light yellow to brown when mature, which are usually white to cream when fresh, turning pale yellow on drying, with bruising brown of pore surface. The tube layer is single or stratified, with pale to purplish brown, hard, and becomes woody when dry. The stipe is central or lateral when present.
The Ganoderma hyphal system is di-trimitic and generative hyphae are thin-walled or occasionally thick-walled, with clamp connections. Skeletal hyphae are hyaline to brown, thick-walled, often long, unbranched. Binding hyphae are almost colourless, thin to thick-walled, branched and with clamp connections. Basidiospores are 7–30 μm long, usually broadly to narrowly ellipsoid, truncate, double-walled, and with an apical germ pore. The endosporium is brown and separated from the hyaline exosporium by inter-wall pillars, negative in Melzer’s reagent (Núñez and Ryvarden 2000; Ryvarden 2004). Basidia are broadly ellipsoid, tapering abruptly at the base, and cystidia are lacking.
Fig. 1 Morphology of Ganoderma species. a An old basidiome of Ganoderma australe, b Mature basidiome of G. casuarinicola, c, d hyphae, e tube layer hyphae, f, g, h Basidiospores, i Pore characteristics. Scale bars: a, b = 2 cm; c, d = 3 μm; e = 15 μm; f, g, h. = 5 μm; i = 500 μm.
Ganoderma species are widely distributed in temperate, subtropical and tropical regions, and appear to thrive in hot and humid conditions (Pilotti et al. 2004; Hapuarachchi et al. 2019a,b,; Luangharn et al. 2019). Basidiomes are commonly in the form of a bracket (Pilotti et al. 2004). Ganoderma is cosmopolitan and an important wood-decaying genus. Some species of Ganoderma are pathogenic, causing root and stem rot on a variety of monocotyledons, dicotyledons and gymnosperms, including a wide range of economically important trees and perennial crops which results in the death of affected trees (Hapuarachchi et al. 2018b). Ganoderma grows as facultative parasites of trees but can also live as saprobes on rotting stumps and roots (Turner 1981; Pilotti et al. 2004). Hence, they have ecological importance in the breakdown of woody plants for nutrient mobilization. Taxa also possess effective machinery of lignocellulose-decomposing enzymes which may be useful for bioenergy production and bioremediation (Hepting 1971; Kües et al. 2015; Hyde et al. 2019).
Several Ganoderma species are prolific sources of highly active bioactive compounds such as polysaccharides, proteins, steroids and triterpenoids. These bioactive compounds show a huge structural and chemical diversity (Shim et al. 2004; Qiao et al. 2005; Wang and Liu 2008; Teng et al. 2011; De Silva et al. 2012a, b; 2013; Hapuarachchi et al. 2017; Li et al. 2018; Hyde et al. 2019). The bioactive constituents have anti-cancer, anti-inflammatory, anti-tumour, anti-oxidant, immunomodulatory, immunodeficiency, anti-diabetic, anti-viral, anti-bacterial, anti-fungal, anti-hypertensive, anti-atherosclerotic, anti-ageing, anti-androgenic, hepatoprotective and radical scavenging properties. They are also promising in neuroprotection, sleep promotion, cholesterol synthesis inhibition, preventing hypoglycemia, inhibition of lipid peroxidation/oxidative DNA damage, maintenance of gut health, prevention of obesity, and stimulation of probiotics (De Silva et al. 2012a; Hapuarachchi et al. 2016a,b, 2017).
Current studies are identifying secondary metabolites, developing models for prediction or early detection of diseases, finding biological control methods as well as understanding genomes. Using artificial neural network spectral analyses and foliage of four disease levels, Ahmadi et al. (2017) provided an early detection method for Ganoderma basal stem rot of oil palm. Sitompul and Nasution (2020) suggested that to control Ganoderma diseases non or weakly pathogenic fungi can be considered as biological control agents. These agents could break down woody debris faster than the pathogen and occupy the same resource as the pathogen (compete for nutrients) as well as producing inhibitory secondary metabolites (Paterson 2007; Sitompul and Nasution 2020). Utomo et al. (2018) sequenced the nuclear genome of G. boninense, the main pathogen of basal stem rot, and the draft genome comprised of 79.24 megabases and 26,226 predicted coding sequences. Ramzi et al. (2019) conducted a study to understand the plant cell wall degradation and pathogenesis of G. boninense via comparative genome analysis. In their study, they found that similarly to G. lucidium, G. boninense was enriched with carbohydrate-active and cell wall degrading enzymes. Following plant-host interaction analysis, several candidate genes including polygalacturonase, endo β-1, 3-xylanase, β-glucanase and laccase were identified as potential cell wall degrading enzymes that contribute to the plant host interaction and pathogenesis. The study provided fundamental knowledge on the fungal genetic ability and capacity to secrete carbohydrate-active and cell wall degrading enzymes. Agudelo-Valencia (2020) pointed out that information regarding the biotechnological importance of Ganoderma species (other than G. lucidium) is quite limited. Therefore, in their study they obtained and studied the genome of G. australe, resulting in gene prediction for the 84-megabase genome, prediction of 22,756 protein-coding genes, prediction of five putative genes and two enzyme complexes from a ganoderic acid pathway.
Most Ganoderma species are pathogenic or facultatively pathogenic, causing root and stem rot on a variety of monocotyledons, dicotyledons, and gymnosperms, including a wide range of economically important trees and perennial crops, which may result in death (Hapuarachchi et al. 2018a). Some species are saprobic and cause white-rot decay of wood (Muthelo 2009). Hence, they have ecological importance in the breakdown of woody plants for nutrient mobilization. They possess effective machinery of lignocellulose-decomposing enzymes useful for bioenergy production and bioremediation (Hepting 1971; Adaskaveg et al. 1991; Kües et al. 2015).
Classification – Basidiomycota, Agaricomycotina, Agaricomycetes, Incertae sedis, Polyporales, Ganodermataceae
Type species – Ganoderma lucidum (Curtis) P. Karst. 1881
Distribution – worldwide
Disease symptoms – basal stem, butt and root rot in economically important trees and perennial crops, especially in tropical regions. Ganoderma disease development is affected by environmental factors and tree death could be either slow or rapid depending on water availability and temperature (Coetzee et al. 2015).
Basal stem rot: Symptoms of basal stem rot disease can take several years to develop, and the presence of the pathogen is often only visible when the fungus is well-established and more than half of the tissue has been decayed. Soils with poor drainage and water stagnation during rainy seasons favour the disease (Kandan et al. 2010).
Butt rot and root rot: The primary symptoms include wilting, mild to severe, of either all leaves or just the lowest leaves in the canopy, premature death of the oldest leaves or a general decline of the tree. The advanced decay of the larger roots is evident after leaves are blown down. Decay may extend from several cms to over a metre into the lower (butt) portion of the tree, depending on the species of Ganoderma. It is quite common for basidiomes not to appear before the severe decline and death of a tree (Elliott and Timothy 2000; Glen et al. 2009). Therefore, the only way to determine if Ganoderma butt rot is the cause is to cut cross-sections through the lower meter or so of the trunk after the tree is felled and examine the cross-sections for the typical pattern of rot: greatest near the soil line, decreasing in sections further from the soil line.
Ganoderma root rot may cause yellowing, wilting, or undersized leaves and dead branches. Tree vigour may decline as the decay of the sapwood advances. The first visible sign of infection is often the formation of basidiomes (solitary or in clusters) on the lower trunk and exposed root areas. There are two types: varnished and unvarnished. The upper surface of varnished fungus rot is typically red-brown with a white edge, shiny, and lacquered. Conks of the unvarnished fungus rot are brown with a white edge weathering to grey (Pilotti et al. 2004). When fresh, both have a white, porous surface on the underside. The rate of decay can lead to death in as little as 3 to 5 years from the time of infection and appears to be determined by tree vigour, which is often influenced by environmental stresses (Nirwan et al. 2016).
Hosts –Ganoderma has a wide host range, with more than 44 species from 34 potential host genera identified (Venkatarayan 1936). The root and stem rots caused by Ganoderma species, result in decreased forestry yields of e.g. Areca catechu (Palanna et al. 2020), Camellia sinensis, Cocos nucifera, (Kinge and Mih 2014), Elaeis guineensis, (Glen et al. 2009) and Hevea brasiliensis (Monkai et al. 2017) worldwide.
Pathogen biology, disease cycle and epidemiology
The fungus is spread by spores produced in the prominent basidiomes that form on the outside of the tree (conks). New spores released from the conks are dispersed throughout the summer during humid periods and infect open wounds on root flares and lower trunk areas of susceptible trees. The spores germinate, and the infection progresses to attack the sapwood of major roots and the lower tree trunk. Over the years, the number of decayed wood increases leading to dangerously soft, spongy wood in the part of the tree that functions as its anchor (Paterson 2007).
Morphology-based identification and diversity
Ganoderma species identification, limitations and their taxonomic segregation have been unclear and recently being debated (Moncalvo et al. 1995; Wang et al. 2009; Cao et al. 2012; Yao et al. 2013; Richter et al. 2015; Zhou et al. 2015). Many Ganoderma collections and species have been misnamed because of the presence of heterogenic forms, taxonomic obstacles and inconsistencies in the way the genus has been subdivided (Mueller et al. 2007). Ganoderma species are genetically heterogeneous, hence a wide range of genetic variation has been reported and caused by outcrossing over generations and different geographical origins (Pilotti et al. 2004). This has led to variation in their listed morphological characteristics, even within the same species (Hong et al. 2001). Environmental factors, variability, inter hybridization and individual morphological bias, mean identification of Ganoderma species is difficult (Zheng et al. 2007a). Hence, naming a species is confused and traditional taxonomic methods based on morphology are inconclusive for establishing a stable classification system for Ganoderma species (Hseu et al. 1996; Hong et al. 2001) which in turn result in an uncertain nomenclature. This confusing situation is mainly the result of various criteria used in identification by different authors. Some authors strictly only focus on host-specificity, geographical distribution and macro morphology of basidiomes, while other authors only focus on spore characteristics as the primarily taxonomic characteristics (Sun et al. 2006; Ekandjo and Chimwamurombe 2012). Richter et al. (2015) suggested using a combination of morphological, chemotaxonomic and molecular methods to develop a more stable taxonomy for this genus.
Molecular identification and diversity
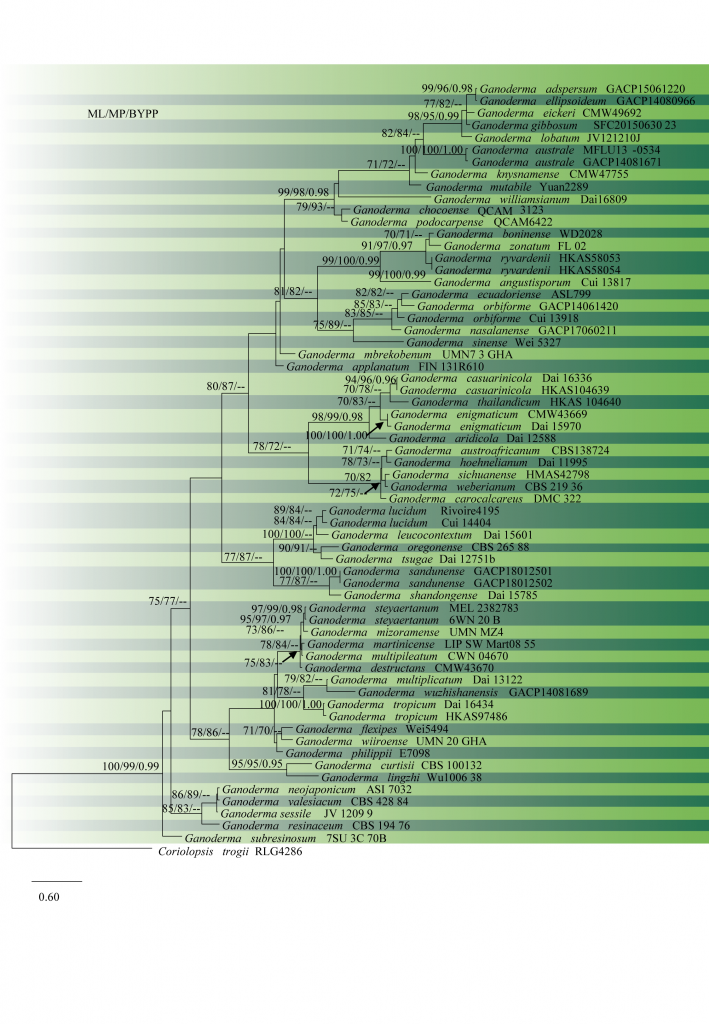
Isoenzyme analysis was the first molecular technique used to separate Ganoderma species (Park et al. 1994; Gottlieb et al. 1995, 1998; Gottlieb and Wright 1999a,b; Smith and Sivasithamparam 2000). Then, DNA sequences of the internal transcribed spacer (ITS), partial large subunit rDNA (Moncalvo et al. 1995, 2000; Cao et al. 2012; Yao et al. 2013; Richter et al. 2015) and nearly complete small subunit rDNA sequences (Hong and Jung 2004; Douanla-Meli and Langer 2009) were used. Later, multigene phylogenetic analyses with protein-coding genes such as β-tubulin (tub2), the largest subunit of RNA polymerase II gene (rpb1), the second-largest subunit of RNA polymerase II (rpb2), and translation elongation factor 1-α (tef1) were performed to resolve the taxonomic confusions within Ganoderma (Park et al. 2012; Zhou et al. 2015; Hennicke et al. 2016; Jargalmaa et al. 2017). However, many problems remain in the resolution of phylogenetic relationships within the genus. As a result of the intricate taxonomy of Ganoderma, 65% of the Ganoderma sequences available in GenBank were reported to be wrongly identified or ambiguously labelled, (Jargalmaa et al. 2017). In this study, we reconstruct the phylogenetic tree based on ITS, tef1 and rpb2 sequence data (Table 1, Fig. 2).
Recommended genetic marker (genus level) – ITS
Recommended genetic markers (species level) – rpb2, tef1
Accepted number of species – There are 456 species and infra-species epithets in Index Fungorum (2020), for 224 accepted species. However, only 64 species have DNA sequence data.
References – Coetzee et al. (2015); Xing et al. (2016, 2018); Tchoumi et al. (2019), Luangharn et al. (2019b), Ye et al. (2019) (phylogeny, new species), Cabarroi-Hernández et al. (2019) (phylogeny).
Table 1 DNA barcodes available for Ganoderma. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Voucher No | ITS | rpb2 | tef1 |
| Ganoderma adspersum | GACP15061220 | MK345425 | MK371437 | MK371431 |
| G. angustisporum | Cui 13817* | MG279170 | MG367507 | MG367563 |
| G. applanatum | FIN131R610 | EF060004 | – | – |
| G. aridicola | Dai 12588* | KU572491 | – | KU572502 |
| G. australe | GACP14081671 | MH106871 | – | – |
| G. australe | MFLU 13-0534 | KP142173 | – | MN423152 |
| G. austroafricanum | CBS138724 | KM507324 | – | – |
| G. boninense | WD 2028 | KJ143905 | KJ143964 | KJ143924 |
| G. carocalcareus | DMC 322* | EU089969 | – | – |
| G. casuarinicola | Dai 16336* | MG279173 | MG367508 | MG367565 |
| G. casuarinicola | HKAS104639 | MK817650 | MK840868 | MK871328 |
| G. chocoense | QCAM3123 | MH890527 | – | – |
| G. curtisii | CBS 100132 | JQ781849 | KJ143967 | KJ143927 |
| G. destructans | CMW43670* | KR183856 | – | – |
| G. ecuadoriense | ASL799 | KU128524 | – | – |
| G. eickeri | CMW49692* | MH571690 | – | MH567287 |
| G. ellipsoideum | GACP14080966* | MH106867 | – | – |
| G. enigmaticum | CBS 139792* | KR183855 | – | – |
| G. enigmaticum | Dai 15970 | KU572486 | MG367513 | KU572496 |
| G. flexipes | Wei 5494 | JN383979 | – | – |
| G. gibbosum | SFC20150630-23 | KY364264 | – | – |
| G. hoehnelianum | Dai11995 | KU219988 | MG367497 | MG367550 |
| G. knysnamense | CMW47755* | MH571690 | – | MH567287 |
| G. leucocontextum | GDGM 40200* | KF011548 | – | – |
| G. lingzhi | Wu 1006-38* | JQ781858 | JX029980 | JX029976 |
| G. lobatum | JV1212/10J | KF605676 | – | KU572501 |
| G. lucidum | Rivoire 4195 | KJ143909 | KJ143969 | – |
| G. lucidum | Cui 14404 | MG279181 | MG367519 | MG367573 |
| G. martinicense | LIP SWMart08-55* | KF963256 | – | – |
| G. mbrekobenum | UMN7-3GHA* | KX000896 | – | – |
| G. mizoramense | UMN-MZ4* | KY643750 | – | – |
| G. multiplicatum | Dai 13122 | KU572488 | – | KU572498 |
| G. multipileum | CWN 04670 | KJ143913 | KJ143972 | KJ143931 |
| G. mutabile | Yuan 2289* | JN383977 | – | – |
| G. nasalanense | GACP17060211* | MK345441 | – | – |
| G. neojaponicum | ASI 7032 | JQ520193 | – | – |
| G. orbiforme | Cui 13918 | MG279186 | MG367522 | MG367576 |
| G. orbiforme | GACP14061420 | MK345447 | – | – |
| G. oregonense | CBS 265.88 | JQ781875 | KJ143974 | KJ143933 |
| G. philippii | E7098 | AJ536662 | – | – |
| G. podocarpense | QCAM6422* | MF796661 | – | – |
| G. resinaceum | CBS 194.76 | KJ143916 | – | KJ143934 |
| G. ryvardenii | HKAS 58053* | HM138671 | – | – |
| G. ryvardenii | HKAS 58054 | HM138672 | – | – |
| G. sandunense | GACP18012501* | MK345450 | – | – |
| G. sandunense | GACP18012502 | MK345451 | – | – |
| G. sessile | JV 1209/9 | KF605629 | – | KJ143936 |
| G. shandongense | Dai 15785 | MG279190 | MG367526 | MG367580 |
| G. sichuanense | HMAS 42798* | JQ781877 | – | – |
| G. sinense | Wei 5327 | KF494998 | MG367529 | KF494976 |
| G. steyaertanum | MEL:2382783 | KP012964 | – | – |
| G. steyaertanum | 6 WN 20B | KJ654462 | – | – |
| G. subresinosum | 7-SU-3-C-70(M)-B | KJ654472 | – | – |
| G. thailandicum | HKAS104640* | MK848681 | MK875831 | MK875829 |
| G. tropicum | Dai 16434 | MG279194 | MG367532 | MG367585 |
| G. tropicum | HKAS 97486 | MH823539 | MH883621 | – |
| G. tsugae | Dai12751b | KJ143919 | KJ143977 | KJ143939 |
| G. valesiacum | CBS428.84 | JQ520218 | – | – |
| G. weberianum | CBS219.36 | JQ520219 | – | – |
| G. wiiroense | UMN-20-GHA | KT952361 | – | – |
| G. williamsianum | Dai 16809 | MG279183 | MG367535 | MG367588 |
| G. wuzhishanensis | GACP14081689 | KU994772 | – | – |
| G. zonatum | FL-02 | KJ143921 | KJ143979 | KJ143941 |
Fig. 2 Phylogram of 64 recognized Ganoderma species, obtained from ML of combined ITS, rpb2, and tef1 datasets. Bootstrap values from ML (left) and MP (middle) greater than 70% and BYPP, greater than 0.95, are indicated above the nodes. The tree is rooted with Coriolopsis trogii. Type specimens are indicated in bold.


No Comments