27 Oct Stemphylium
Stemphylium Wallr., Flora Cryptogamica Germaniae 2: 300 (1833)
Background
Stemphylium mainly comprises saprobes or weak plant pathogens (Woudenberg et al. 2017). However, some species are primary pathogens causing leaf blight on various crops, resulting in yield and economic losses (Hanse et al. 2015; Brahmanage et al. 2018). The asexual morph is a dematiaceous hyphomycete while the sexual morph was previously defined as Pleospora sensu stricto (Inderbitzin et al. 2009; Woudenberg et al. 2017). Rossman et al. (2015) recommended the use of Stemphylium over Pleospora which has been followed by various authors (Hongsanan et al. 2017, 2020; Wijayawardene et al. 2018, 2020). Stemphylium is one of the most important moulds human allergens in the USA (Gutiérrez-Rodríguez et al. 2011). Brahmanage et al. (2018) discussed the pathogenicity, disease severity, distribution and molecular phylogenetic affinities of pathogenic isolates of Stemphylium.
Stemphylium leaf blight caused by S. versicarum was identified as an emerging disease in New York, USA. Sharma et al. (2020b) provided two genome resources for two S. versicarum isolates from leaf blight of onion. Genomic data allows for an understanding of the population biology, fungicide resistance, as well as development of control strategies against the disease. Pathogenesis related 511 secreted proteins were predicted from S. lycopersici by Zeng et al. (2018) which helps in understanding the roles of proteins in host penetration and tissue necrosis. Stemphylium loti secretes Tenuazonic acid, inhibiting the plant plasma membrane H+-ATPase, which results in membrane potential depolarization and eventually necrosis (Bjørk et al. 2019). Su et al. (2019) fine-mapped the tomato grey spot resistance gene Sm, in a 185kb region through a map-based cloning strategy. Leach et al. (2020) identified a relationship between thrips (Thrips tabaci) and S. vesicarium in the development of Stemphylium leaf blight in onion.
Classification – Ascomycota, Pezizomycotina, Dothideomycetes, Pleosporomycetidae, Pleosporales, Pleosporaceae
Type species – Stemphylium botryosum Wallr.
Distribution – worldwide
Disease symptoms – Gray spot, Stemphylium leaf blight (Leaf spot, defoliation, curling and bending of the leaf margins and stems)
Initial symptoms of the leaves are small, irregular, brown spots. Generally, the spots gradually lighten and eventually become greyish as they become necrotic and dry. When severe, yellow spots can be seen throughout all leaves of the plant and the heavily infected leaves die (Basallote-Ureba et al. 1999, Crous et al. 2016; Brahamanage et al. 2018).
Hosts – Species are pathogenic on a wide range of hosts including Amaryllidaceae, Asparagaceae, Fabaceae, Malvaceae, Poaceae, Rosaceae and Solanaceae
Pathogen biology, disease cycle and epidemiology
Species can survive as saprobes on crop residues, soil, plant debris and on many alternative hosts and ascospores become the primary inocula in the following season. Once the disease is established during favourable conditions, conidial production in primary lesions may occur, dispersing spores to healthy plants by wind and rain splashing. Environmental factors such as temperature and moisture are key factors in disease development. Seedlings of plants can transmit the diseases if they become infected in the nursery (Basallote-Ureba et al. 1998, 1999; Boshuizen et al. 2004; Zheng et al. 2010; Blancard 2012). However, to date, diseases and epidemiology such as factors affecting the disease development, interactions with different hosts and genetics of host resistance are poorly studied (Das et al. 2019).
Morphological based identification and diversity
Species can be distinguished from other hyphomycetes in Pleosporaceae forming phaeodictyospores, based on percurrent proliferation of its conidiophores and apically swollen conidiogenous cells (Köhl et al. 2009). Simmons (1967) established criteria for morphological identification of various Stemphylium species and introduced Pleospora herbarum as the sexual morph of the type species Stemphylium botryosum. However, Simmons (1985) subsequently reclassified and reported Pleospora tarda as the sexual morph of Stemphylium botryosum and Pleospora herbarum as the sexual morph of Stemphylium herbarum (Moslemi et al. 2017). Morphological features, such as size and time of pseudothecial maturation, conidiophores and conidia and ascospore shape and size can be considered as important characteristics in species identification (Câmara et al. 2002; Fig. 1).
Köhl et al. (2009) and Woudenberg et al. (2017) pointed out that the lack of (ex-) type material of species and morphology-based species identifications without molecular evidence make it difficult in determining correct species nomenclature. Therefore, relying on morphological characters alone in identifying species is not recommended.
Fig. 1 Stemphylium sp. a. Ascomata on host b. Vertical section through an ascoma c. immature and mature asci d. Pseudoparaphyses e. Ascospores f. Ascospores in Indian ink. Scale bars: b = 50 μm, c-f = 10 μm.
Molecular based identification and diversity
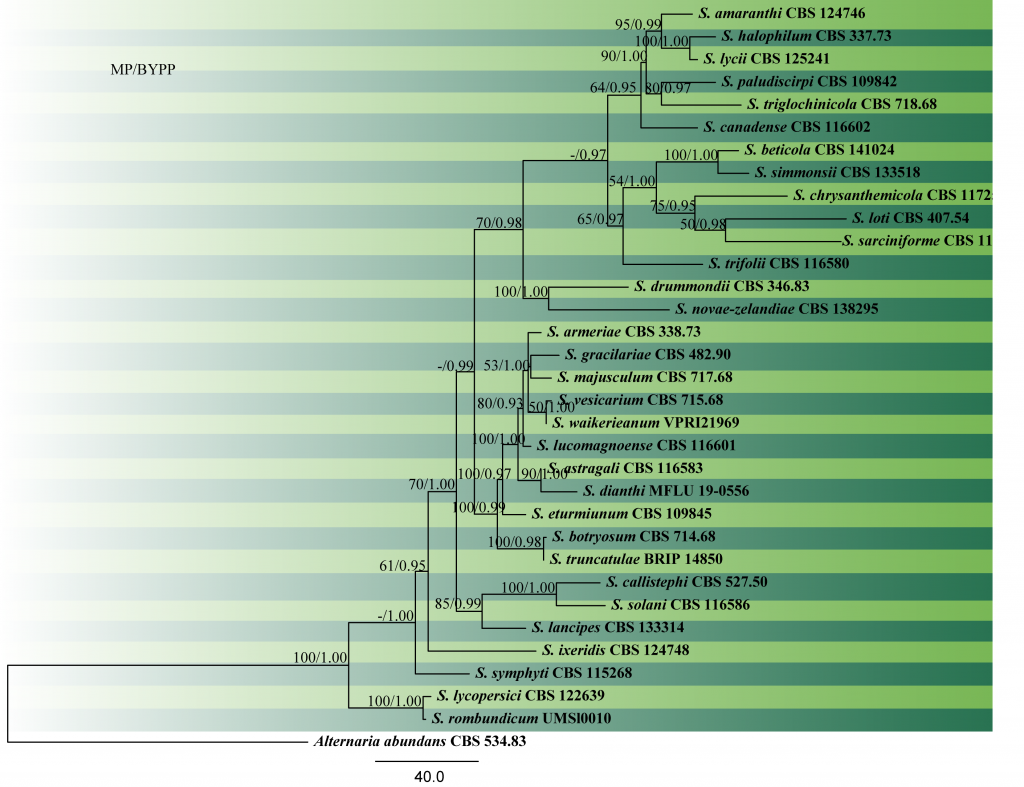
ITS (rDNA) and glyceraldehyde-3-phosphate dehydrogenase (gapdh) sequences were used by Câmara et al. (2002) to confirm the monophyly of Stemphylium. In the extensive study of 110 Stemphylium strains from various hosts and DNA sequence data of ITS, gapdh and tef1 loci and the intergenic spacer between vmaA and vpsA, Inderbitzin et al. (2009) identified 23 representatives derived from type strains, while 40 strains remained unnamed. Woudenberg et al. (2017) revised the genus and accepted 28 species, synonymizing 22 names and proposing two new combinations based on combined analyses of the ITS, gapdh and cmdA gene regions. Marin-Felix et al. (2019) introduced three new species (S. rombundicum, S. truncatulae and S. waikerieanum), while Brahmanage et al. (2018) introduced S. dianthi based on multi loci phylogeny. In this study, we reconstruct the phylogeny based on combined ITS, gapdh and cmdA sequence data (Fig. 2).
Recommended genetic marker (genus level) – ITS
Recommended genetic markers (species level) – cmdA, gapdh
Accepted number of species – There are 207 epithets listed in Index Fungorum, however only 32 species have DNA sequence data (Table 1).
References –Simmons 1967, Köhl et al. 2009 (morphology); Câmara et al. 2002, Inderbitzin et al. 2009, Moslemi et al. 2017, Woudenberg et al. 2017, Brahmanage et al. 2019, Marin-Felix et al. 2019 (morphology and phylogeny)
Table 1 DNA barcodes available for Stemphylium. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species name | Isolate/specimen no | ITS | gapdh | cmdA |
| Stemphylium amaranthi | CBS 124746* | KU850505 | KU850652 | KU850793 |
| S. armeriae | CBS 338.73 | KU850511 | KU850658 | KU850799 |
| S. astragali # | CBS 116583* | KU850512 | KU850659 | KU850800 |
| S. beticola# | CBS 141024* | KU850520 | KU850667 | KU850808 |
| S. botryosum# | CBS 714.68* | KC584238 | AF443881 | KU850826 |
| S. callistephi | CBS 527.50* | KU850539 | KU850686 | KU850828 |
| S. canadense | CBS 116602* | KU850641 | KU850782 | KU850932 |
| S. chrysanthemicola# | CBS 117255* | KU850640 | KU850781 | KU850931 |
| S. dianthi | MFLU 19-0556* | MK500718 | – | MK500734 |
| S. drummondii | CBS 346.83* | GQ395365 | KU850687 | KU850829 |
| S. eturmiunum# | CBS 109845* | KU850541 | KU850689 | KU850831 |
| S. gracilariae | CBS 482.90* | KU850549 | AF443883 | KU850839 |
| S. halophilum | CBS 337.73* | KU850553 | KU850700 | KU850843 |
| S. ixeridis | CBS 124748* | KU850590 | KU850737 | KU850881 |
| S. lancipes | CBS 133314* | KU850596 | KU850742 | KU850887 |
| S. loti | CBS 407.54* | KU850597 | KU850743 | KU850888 |
| S. lucomagnoense | CBS 116601* | KU850629 | KU850770 | KU850920 |
| S. lycii | CBS 125241* | KU850602 | KU850748 | KU850893 |
| S. lycopersici | CBS 122639* | KU850611 | KU850756 | KU850902 |
| S. majusculum | CBS 717.68* | KU850618 | AF443891 | KU850909 |
| S. novae-zelandiae | CBS 138295* | KU850631 | KU850772 | KU850922 |
| S. paludiscirpi | CBS 109842* | KU850620 | KU850762 | KU850911 |
| S. rombundicum | BRIP 27486* | MK336819 | MK336865 | MK336842 |
| S. sarciniforme | CBS 110049* | KU850591 | KU850738 | KU850882 |
| S. simmonsii# | CBS 133518* | KU850637 | KU850778 | KU850928 |
| S. solani# | CBS 116586* | KU850627 | KU850768 | KU850918 |
| S. symphyti | CBS 115268* | KU850643 | KU850784 | KU850934 |
| S. trifolii | CBS 116580* | KU850647 | KU850788 | KU850938 |
| S. triglochinicola | CBS 718.68* | KU850648 | KU850789 | KU850939 |
| S. truncatulae | BRIP 14850* | MK336815 | MK336861 | MK336838 |
| S. vesicarium# | CBS 715.68* | KU850565 | KU850712 | KU850855 |
| S. waikerieanum | VPRI 21969* | MK336832 | MK336878 | MK336855 |
Fig. 2 Phylogram generated from MP analysis based on combined sequences of ITS, gapdh and cmdA sequences of all species of Stemphylium. Related sequences were obtained from GenBank. Thirty three taxa are included in the analyses, which comprise 1936 characters including gaps, of which 1355 characters are constant, 271characters are parsimony-uninformative and 310 characters parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of four equally most parsimonious trees with a length of 1112 steps (CI = 0.660, RI=0.721, RC = 0.476, HI = 0.340) in the first tree. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Alternaria abundance (CBS 534.83). MP bootstrap support value ≥50% and BYPP ≥0.9 are shown respectively near the nodes. Ex-type strains are in bold.


No Comments