26 Oct Neoerysiphe
Neoerysiphe U. Braun, Schlechtendalia 3: 50 (1999)
Background
Neoerysiphe was classified in section Galeopsidis within Erysiphe. Phylogenetic analysis, however, showed Erysiphe to be polyphyletic, and Galeopsidis was raised to generic rank (Takamatsu et al. 1998; Braun 1999; Saenz and Taylor 1999). Therefore, in the current classification Neoerysiphe belongs to the tribe Golovinomyceteae.
Classification – Erysiphaceae, Erysiphales, Leotiomycetidae, Leotiomycetes, Pezizomycotina
Type species – Neoerysiphe galeopsidis (DC.) U. Braun
Distribution – Argentina, Australia, Belarus, Brazil, Bulgaria, Canada, China, Denmark, Finland, France, Germany, Hungary, India, Israel, Italy, Japan, Korea, Netherlands, Norway, Poland, Romania, Russia, Slovakia, Sweden, Switzerland, Turkey, UK, Ukraine and USA (Farr and Rossman 2020).
Disease symptoms-powdery mildew
Hosts – Neoerysiphe species have a wide host distribution infecting more than 300 species from families including Asteraceae, Acanthaceae, Bignoniaceae, Elaeocarpaceae, Lamiaceae, Rubiaceae and Verbenaceae (Amano 1986; Braun 1999; Bahcecioglu et al. 2006). In general, all species have a specific host range confined to one plant family, except N. galeopsidis which affects several species in four families (Takamatsu et al. 2008).
Pathogen biology, disease cycle and epidemiology
Discussed under Erysiphaceae.
Morphological based identification and diversity
Neoerysiphe is in the tribe Golovinomyceteae with Arthrocladiella and Golovinomyces. These genera share a common asexual morph characterized by catenate conidia without distinct fibrosin bodies (Braun 1999). Neoerysiphe is characterized by lobed appressoria and the striate surface of the conidia (Braun 1981; Cook et al. 1997; Braun and Cook 2012). Braun and Cook (2012) mentioned that 15 species of Neoerysiphe are described on different hosts belonging to 11 plant families. Of these 15 species, 11 sexual morphs and 14 asexual morphs have been identified (except N. joerstadii) (Heluta et al. 2010; Braun and Cook 2012). Striatodium is now considered as a synonym of Neoerysiphe and three species viz. N. aloysiae, N. baccharidis and N. maquii were transferred to Neoerysiphe, while Striatodium jaborosae was not transferred as its phylogenetic position are unclear (Wijayawardene et al. 2017a).
Molecular based identification and diversity
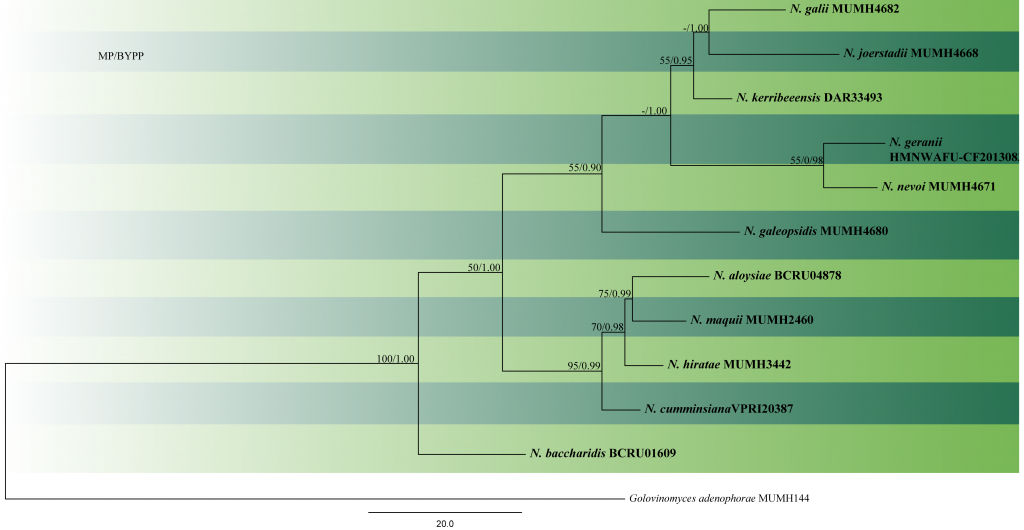
The phylogenetic placement of Neoerysiphe within Erysiphaceae has been reported in a few papers (Saenz and Taylor 1999; Mori et al. 2000; Cook et al. 2006). However, these treatments used only limited sequence data for the genus. Takamatsu et al. (2008) conducted the first comprehensive study on this genus using ITS sequence data and the divergent domains D1 and D2 of the 28S rDNA for 30 strains. In their study, the 30 taxa, clustered into three monophyletic groups that were represented by N. galeopsidis on Lamiaceae, N. galii on Rubiaceae and N. cumminsiana from Asteraceae. Takamatsu et al. (2008) used an LSU dataset to estimate the timing of divergence of Neoerysiphe. Neoerysiphe split from other genera ca 35–45 Mya and the three groups of Neoerysiphe diverged between 10 and 15 Mya in the Miocene. Heluta et al. (2010) used 65 ITS sequences in their analyses for identifying Neoerysiphe species infecting Asteraceae and Geranium in Eurasia and introduced three new species, viz. N. hiratae, N. joerstadii and N. nevoi. Gregorio-Cipriano et al. (2020) introduced a new species N. sechii causing powdery mildew on Sechium edule and S. mexicanum in Mexico. The authors mentioned that they were unable to recover DNA in pure form from some samples, as fragments of infected leaves were used during the extraction. Therefore, a specific oligonucleotide for Erysiphales at the 5= region of ITS was designed: ErysiF (5=-AGGATCATTACWGAGYGYGAG-3=) was used along with NLP1 (Limkaisang et al. 2006) to amplify a fragment of approximately 1200 bp (that included the ITS1-5.8S-ITS2 region and a section of approximately 680 nucleotides from 28S). Species used in the phylogenetic analyses done in this study are listed in Table 1 and given in Fig. 1.
Recommended genetic marker (genus level) – ITS and LSU
Recommended genetic markers (species level) – ITS
Accepted number of species – There are 16 species epithets in Index Fungorum (2020), for 15 accepted species. However, only 12 species have DNA sequence data (N. chelones, N. gnaphalii and N. rubiae do not have molecular data) (Table 1).
References – Takamatsu et al. 1998, Braun 1999, Saenz and Taylor 1999 (morphology); Heluta et al. 2010, Braun and Cook 2012, Gregorio-Cipriano et al. 2020 (morphology and phylogeny).
Table 1 DNA barcodes available for Neoerysiphe. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains and voucher strains are in bold.
| Species | Strain no | ITS | LSU |
| Neoerysiphe aloysiae | BCRU 04878 | AB329683 | – |
| N. baccharidis | BCRU 01609 | AB329685 | AB329684 |
| N. cumminsiana | VPRI 20387 | GU356539 | – |
| N. galeopsidis | MUMH 4680 | AB498949 | AB022369 |
| N. galii | MUMH 4682 | AB498951 | AB103365 |
| N. geranii | HMNWAFU-CF2013083 | KR048092 | KR048161 |
| N. hiratae | MUMH 3442 | AB498962 | – |
| N. joerstadii | MUMH 4668 | AB498976 | – |
| N. kerribeeensis | DAR 33493 | GU356546 | – |
| N. maquii | MUMH 2460 | AB329686 | – |
| N. nevoi | MUMH 4671 | AB498975 | – |
Fig. 1 Phylogram generated from MP analysis based on combined sequences of ITS and LSU sequences of all species of Neoerysiphe with molecular data. Related sequences were obtained from GenBank. 12 taxa are included in the analyses, which comprise 2023 characters including gaps, of which 1790 characters are constant, 167characters are parsimony-uninformative and 66 characters parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of four equally most parsimonious trees with a length of 316 steps (CI = 0.848, RI=0.678, RC = 0.575, HI = 0.152) in the first tree. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Golovinomyces adenophorae (MUMH144). MP bootstrap support value ≥50% and BYPP ≥0.9 are shown respectively near the nodes. Ex-type strains are in bold.

No Comments