27 Oct Colletotrichum
Colletotrichum Corda, in Sturm, Deutschl. Fl., 3 Abt. (Pilze Deutschl.) 3(12): 41 (1831)
Background
Colletotrichum was introduced by Corda (1831), belonging to Glomerellaceae (Glomerellales, Sordariomycetes), and is the sole member of this family (Maharachchikumbura et al. 2015, 2016; Hyde et al. 2020b). Species may occupy different lifestyles, ranging from necrotrophy to hemibiotrophy as well as endophytism (Crouch et al. 2014). Colletotrichum species are important plant pathogens in both tropical and temperate regions on many economically important crops (Hyde et al. 2009a,b, 2014; Cannon et al. 2012; Jayawardena et al. 2016b,c). Based on recognized scientific and economic importance this genus was voted the eighth most important plant pathogenic group in the world (Dean et al. 2012). Colletotrichum species have been identified as endophytes (Manamgoda et al. 2013; Tao et al. 2013; Hyde et al. 2014; Jayawardena et al. 2016c) and some are saprobes on dead plant material (Photita et al. 2005; Jayawardena et al. 2016b). A few species have been identified to be pathogenic to humans (C. coccodes, C. dematium, C. gloeosporioides (Natarajan et al. 2013)) and on insects (C. fioriniae (Damm et al. 2012b)). Colletotrichum species are cosmopolitan in distribution and show a diverse hosts association (Sharma et al. 2015). A host plant genus can be infected by many Colletotrichum species (Silva et al. 2012; Jayawardena et al. 2016c), and on the contrary, a single species of Colletotrichum can infect many host plants (Damm et al. 2012a, b; Weir et al. 2012).
Correct species identification is important to understand the species diversity, plant pathology and quarantine, concerning human infections, agriculture, bio-control, plant breeding, whole-genome sequencing, developing and maintaining knowledge databases, bioprospecting and understanding the evolutionary history (Jayawardena et al. 2016a). Due to a small number of distinctive morphological characters available for identification, misidentification of these species is frequent. Misapplication and misidentification of species are also due to the misunderstanding of their host-specific nature, ambiguous species boundaries and incorrect sequences (Cannon et al. 2012; Hyde et al. 2014; Jayawardena et al. 2016a). Therefore, having a stable taxonomy for the identification of these species is a significant practical concern (Shenoy et al. 2007). To establish a natural classification system, researchers strongly recommend the use of geographical, ecological, morphological and genetic data (Cai et al. 2009; Sharma and Shenoy 2016).
Species of Colletotrichum are extensively studied as model organisms (Cannon et al. 2012; Hyde et al. 2014). This enables the researchers to understand the pathogen variation, infection mechanism, evolution and population dynamics. Pathogenicity genes of C. graminicola, C. higginsianum and C. orbiculare have been studied (Huser et al. 2009; O’Connell et al. 2012). Asakura et al. (2009) discovered the importance of the pexophagy factor ATG26 for appressorium formation. A total of 28 genome projects that include 25 different Colletotrichum species can be found; 15 of these strains are still at the annotation stage and 13 are now at the ‘Fungal Standard Draft’ stage (Carbú et al. 2019). These genomes will allow further analysis of species diversity and evolutionary mechanisms and may serve as a foundation for genetic analysis that leads to a greater understanding of interactions between plants and fungal pathogens (Meng et al. 2020). Baroncelli et al. (2016) studied four strains of C. acutatum and illustrated the plasticity of Colletotrichum genomes and showed that major changes in host range are associated with relatively recent changes in gene content. A genome of C. fructicola from apple in China was compared with its reference genome, which identified a number of strong duplication/loss events at key phylogenetic nodes (Liang et al. 2018). Gan et al. (2019) provided the updated genome for C. orbiculare and also provided three draft genomes for C. trifolli, C. sidae and C. spinosum. Colletotrichum higginsianum has a compartmentalized genome consisting of gene-sparse, transposable elements dense regions with more effector candidate genes and gene-dense, TE-sparse regions harbouring conserved genes which help the pathogen to generate genomic diversity (Tsushima et al. 2019). Comparative genome analysis indicated that there is a rapid evolution of pathogenicity genes in C. tanaceti (Lelwala et al. 2019).
Species of Colletotrichum can be used as biocontrol agents and as biocatalysts (C. dematium, C. gloeosporioides, C. graminicola, C. lindemuthianum, C. orbiculare, C.theobromicola, C. trifoli (Jayawardena et al. 2016b)). Jayawardena et al. (2016b) discussed the importance of secondary metabolites produced by species with relation to pathogenesis, medicines, disease control and toxins.
Classification – Ascomycota, Pezizomycotina Sordariomycetes, Hypocreomycetidae, Glomerellales, Glomerellaceae
Type species – Colletotrichum lineola Corda, in Sturm, Deutschl. Fl., 3 Abt. (Pilze Deutschl.) 3(12): 41 (1831)
Distribution – Worldwide
Disease symptoms – Anthracnose disease, red rot, crown and stem rots, ripe rot, seedling blights and brown blotch.
Anthracnose disease symptoms include defined, often sunken necrotic spots on leaves, stems, flowers or fruits and may show a lot of variation depending on the host (Fig 1a−e).
Hosts – Pathogens on many host families including, Amaryllidaceae, Amaranthaceae, Anacardiaceae, Annonaceae, Apiaceae, Apocynaceae, Araceae, Araliaceae, Arecaceae, Asparagaceae, Asteraceae, Bignoniaceae, Campanulaceae, Caricaceae, Crassulaceae, Cucurbitaceae, Euphorbiaceae, Fabaceae, Iridaceae, Lamiaceae, Lauraceae, Malvaceae, Melastomataceae, Menispermaceae, Moraceae, Myrtaceae, Oleaceae, Olivaceae, Orchidaceae, Passifloraceae, Pinaceae, Piperaceae, Plumbaginaceae, Poaceae, Podocarpaceae, Polemoniaceae, Proteaceae, Ranunculaceae, Rosaceae, Rubiaceae, Rutaceae, Solanaceae, Theaceae and Vitaceae.
Pathogen biology, disease cycle and epidemiology
For Colletotrichum biology, disease cycle and epidemiology see Cannon et al. (2012) and De Silva et al. (2017).
Morphological based identification and diversity
Due to the overlapping morphological characters, species delimitation based on morphology alone is hardly possible (Jayawardena et al. 2016b; Marin-Felix et al. 2017; Fig 1 f-l).
Fig. 1 Colletotrichum sp. a. Chilli anthracnose symptoms b. Crown rot of strawberry c. Leaf blight of Cannas sp. d-e strawberry anthracnose symptoms f-g Conidiomata and spore mass h Conidiophore i Setae j Conidiogenous cells and curved conidia k Conidia of C. gloeosporioides l Appressoria
Molecular based identification and diversity
Cai et al. (2009) proposed the use of a polyphasic approach with multi-loci sequence analyses combined with geographical, ecological and morphological data for reliable species delimitation. Application of this polyphasic approach resulted in the delimitation of almost 200 species, most of them belonging to species complexes such as acutatum, boninense and gloeosporioides. There is no universal set of loci to use when identifying Colletotrichum species. Cannon et al. (2012), Damm et al. (2012a,b, 2013, 2014, 2019, Liu et al. 2016) used ITS, gapdh, chs, act, his and tub2 (with some also gs or cal) for studying species within the acutatum, boninense, dematium, destructivum, gigasporum, orbiculare, spaethianum and truncatum species complexes, while Weir et al. (2012) additionally applied gs, cal and sod2 within the gloeosporioides species complex. Hyde et al. (2014), Jayawardena et al. (2016b), Marin-Felix et al. (2017) used ITS, gapdh, chs, act and tub2 to differentiate the species. Using five loci for the whole genus gave similar results to 6-7 loci used for the whole genus. In contrast, Crouch et al. (2009b) applied ITS, sod2, apn2 and mat1/apn2, to study the graminicola and caudatum species complexes. Use of ApMat locus to delimit the species within gloeosporioides species complex was emphasized by Silva et al. (2012) and Sharma et al. (2015) as it provides a higher resolution when compared to previously used loci. However, studies by Liu et al. (2015, 2016) revealed that using this locus with other loci would provide a satisfactory species delimitation within the gloeosporioides species complex. For species delimitation, application of the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) has proven to be a powerful tool (Cai et al. 2009). Coalescent-based species delimitation methods can also be used to infer the dynamic of divergence, evolutionary process and the relationships among species (McCormack et al. 2009, Liu et al. 2016).
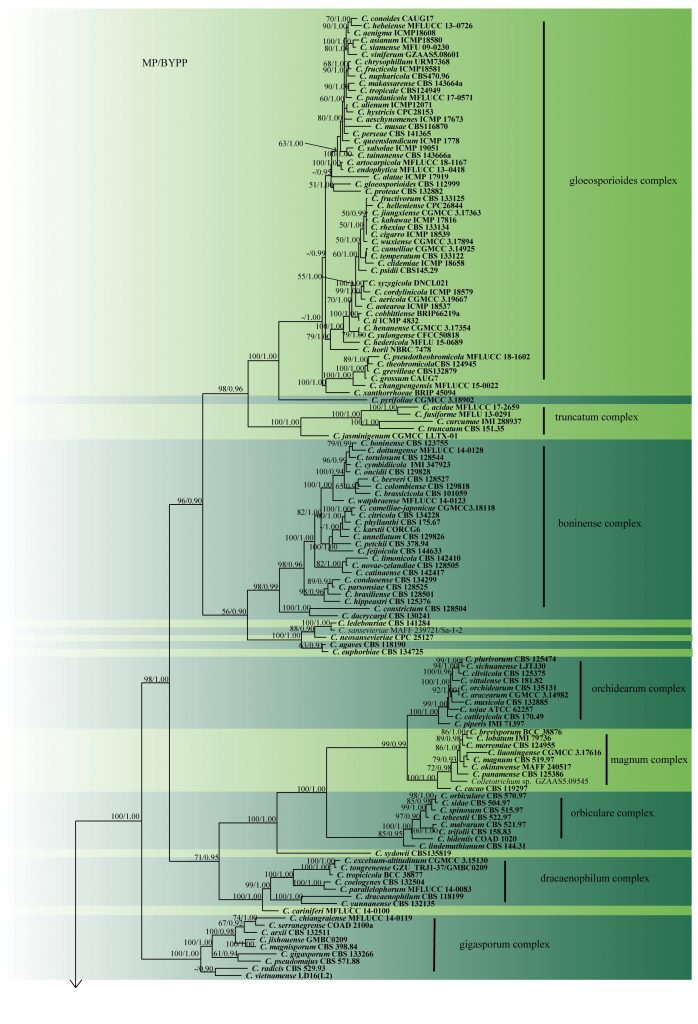
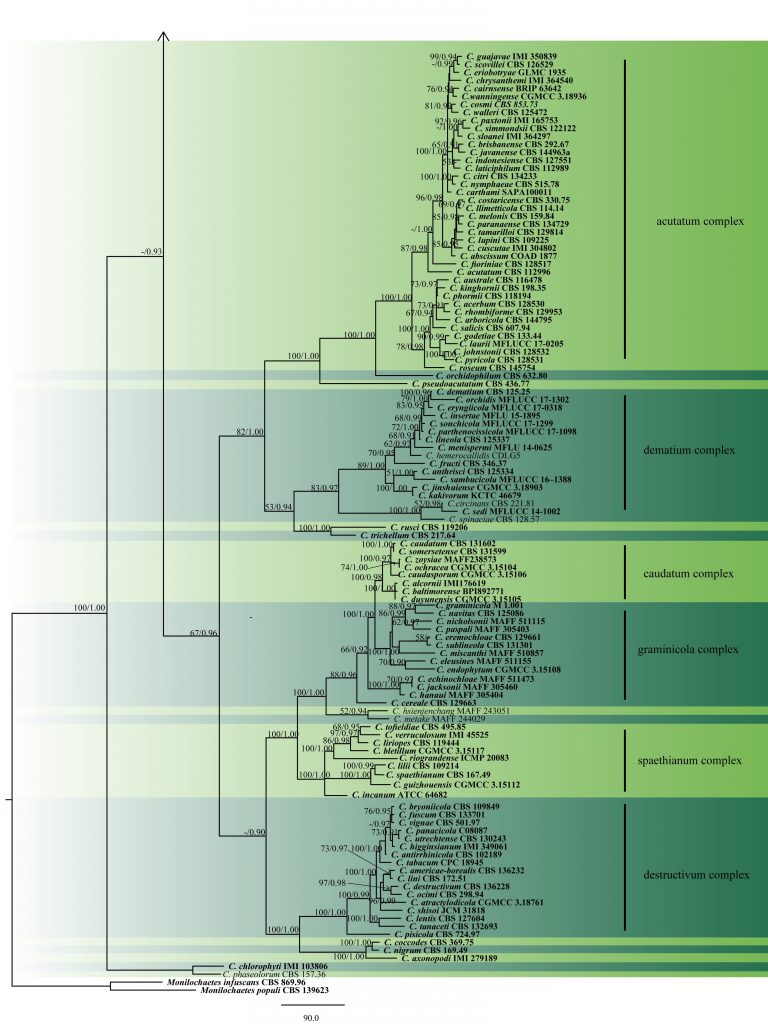
Jayawardena et al. (2016b) provided the accepted species for the genus with backbone trees for each species complex and with notes for each accepted species. De Silva et al. (2017) reviewed the lifestyles and how this can be applied to plant biosecurity. Ariyawansa et al. (2015), Yan et al. (2015), Li et al. (2016), De Silva et al. (2017), Hyde et al. (2017, 2020a, b), Marin-Felix et al. (2017), Tibpromma et al. (2017, 2018), Samarakoon et al. (2018), Bhunjun et al. (2019) have introduced new species based on morphology, phylogeny and GCPSR evidence. Damm et al. (2019) introduced three new species complexes namely, dracaenophilum, magnum and orchidearum based on morphology and phylogeny. Cabral et al. (2020) based on pathological, morphological, cytogenomic, biochemical and molecular data, assigned the previously known C. kahawae subsp. ciggario as a new species, C. ciggario. At present, based on multi-loci phylogeny there are 14 species complexes. With 59 new species having been added since the last treatment (Marin-Felix et al. 2017), here we present an analysis using five loci (Table 1) for all Colletotrichum species (Fig. 2). From the studies conducted on this genus, it is clear that the resolution of species differs depending on both locus and species. Therefore, to select a better genetic marker and the best secondary barcoding gene/genes is still an ongoing process.
Recommended genetic marker (genus level) – ITS
Recommended genetic markers (species level) – act, apmat, apn2, cal, chs-1, gapdh, gs, his, mat1/apn2, sod2, and tub2.
Accepted number of species – There are 903 epithets listed in Index Fungorum (2020), however, 247 species with molecular data are treated as accepted (Table 1).
References – Cai et al. 2009 (polyphasic approach); Hyde et al. 2009a,b (morphology and accepted species); Cannon et al. 2012 (A review and an updated account of the genus); Crouch 2009a,b,c, 2014, Damm et al. 2009, 2012a,b, 2013, 2014, 2019, Weir et al. 2012 (morphology and phylogeny); Hyde et al. 2014, Jayawardena et al. 2016b, Marin-Felix et al. 2017 (accepted number of species).
Table 1 DNA barcodes available for Colletotrichum, isolates used in the phylogenetic analyses. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains in bold and marked with an asterisk (*).Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species name | Isolate No | ITS | gapdh | chs-1 | act | tub2 | Complex |
| Colletotrichum abscissum# | COAD 1877* | KP843126 | KP843129 | KP843132 | KP843141 | KP843135 | acutatum |
| C. acerbum | CBS 128530* | JQ948459 | JQ948790 | JQ949120 | JQ949780 | JQ950110 | acutatum |
| C. acidae | MFLUCC 17-2659* | MG996505 | MH003691 | MH003694 | MH003697 | MH003700 | truncatum |
| C. acutatum# | CBS 112996* | JQ005776 | JQ948677 | JQ005797 | JQ005839 | JQ005860 | acutatum |
| C. aenigma# | ICMP 18608* | JX010244 | JX010044 | JX009774 | JX009443 | JX010389 | gloeosporioides |
| C. aericola# | CGMCC 3.19667* | MK914635 | MK935455 | MK935541 | MK935374 | MK935498 | gloeosporioides |
| C. aeschynomenes# | ICMP 17673 | JX010176 | JX009930 | JX009799 | JX009483 | JX010392 | gloeosporioides |
| C. agaves | CBS 118190* | DQ286221 | singleton | ||||
| C. alcornii | IMI176619* | JX076858 | caudatum | ||||
| C. alatae# | CBS 304.67* | JX010190 | JX009990 | JX009837 | JX009471 | JX010383 | gloeosporioides |
| C. alienum# | ICMP 12071* | JX010251 | JX010028 | JX009882 | JX009572 | JX010411 | gloeosporioides |
| C. americae-borealis# | CBS 136232* | KM105224 | KM105579 | KM105294 | KM105434 | KM105504 | destructivum |
| C. annellatum | CBS 129826* | JQ005222 | JQ005309 | JQ005396 | JQ005570 | JQ005656 | boninense |
| C. anthrisci | CBS 125334* | GU227845 | GU228237 | GU228335 | GU227943 | GU228139 | dematium |
| C. antirrhinicola | CBS 102189* | KM105180 | KM105531 | KM105250 | KM105390 | KM105460 | destructivum |
| C. aotearoa | ICMP 18537* | JX010205 | JX010005 | JX009853 | JX009854 | JX010420 | gloeosporioides |
| C. araceaerum | CGMCC 3.14982* | KX853166 | KX893585 | KX893577 | KX893581 | singleton | |
| C. arboricola# | CBS 144795* | MH817944 | MH817950 | MH817956 | MH817962 | acutatum | |
| C. artocarpicola# | MFLUCC 18-1167* | MN415991 | MN435568 | MN435569 | MN435570 | MN435567 | gloeosporioides |
| C. arxii | CBS 132511* | KF687716 | KF687843 | KF687780 | KF687802 | KF687881 | gigasporum |
| C. asianum# | ICMP 18580* | JX010196 | JX010053 | JX009867 | JX009584 | JX010406 | gloeosporioides |
| C. atractylodicola | CGMCC 3.18761* | KR149280 | KR259334 | KR259333 | KR132243 | KU058178 | destructivum |
| C. australe | CBS 116478* | JQ948455 | JQ948786 | JQ949116 | JQ949776 | JQ950106 | acutatum |
| C. axonopodi | IMI 279189/279189AA | EU554086 | singleton | ||||
| C. beeveri | CBS 128527* | JQ005171 | JQ005258 | JQ005345 | JQ005519 | JQ005605 | boninense |
| C. baltimorense | BPI892771* | JX076866 | caudatum | ||||
| C.bidentis | COAD 1020* | KF178481 | KF178506 | KF178530 | KF178578 | KF178602 | orbiculare |
| C. bletillum | CGMCC 3.15117* | JX625178 | KC843506 | KC843542 | JX625207 | spaethianum | |
| C. boninense# | CBS 123755* | JQ005153 | JQ005240 | JQ005327 | JQ005501 | JQ005588 | boninense |
| C. brasiliense | CBS 128501* | JQ005235 | JQ005322 | JQ005409 | JQ005583 | JQ005669 | boninense |
| C. brassicicola | CBS 101059* | JQ005172 | JQ005259 | JQ005346 | JQ005520 | JQ005606 | boninense |
| C. brevisporum# | BCC 38876* | JN050238 | JN050227 | JN050216 | JN050244 | magnum | |
| C. brisbaniense | CBS 292.67* | JQ948291 | JQ948621 | JQ948952 | JQ949612 | JQ949942 | acutatum |
| C. bryoniicola | CBS 109849* | KM105181 | KM105532 | KM105251 | KM105391 | KM105461 | destructivum |
| C. cacao# | CBS 119297* | MG600772 | MG600832 | MG600878 | MG600976 | MG601039 | magnum |
| C. cairnsense# | BRIP 63642* | KU923672 | KU923704 | KU923710 | KU923716 | KU923688 | acutatum |
| C. camelliae# | CGMCC3.14925* | KJ955081 | KJ954782 | KJ954363 | KJ955230 | gloeosporioides | |
| C. camelliae-japonicae | CGMCC3.18118* | KX853165 | KX893584 | KX893576 | KX893580 | boninense | |
| C. carthami# | SAPA100011* | AB696998 | AB696992 | acutatum | |||
| C. catinaense# | CBS 142417* | KY856400 | KY856224 | KY856136 | KY855971 | KY856482 | boninense |
| C. cattleyicola | CBS 170.49* | MG600758 | MG600819 | MG600866 | MG600963 | MG601025 | orchidearum |
| C. cariniferi | MFLUCC 14-0100* | MF448521 | MH351274 | singleton | |||
| C. caudatum | CBS 131602* | JX076860 | caudatum | ||||
| C. caudasporum | CGMCC 3.15106* | JX625162 | KC843512 | KC843526 | JX625190 | caudatum | |
| C. cereale# | CBS 129663* | JQ005774 | JQ005795 | JQ005837 | JQ005858 | graminicola | |
| C. changpingense# | MFLUCC 15-0022* | KP683152 | KP852469 | KP852449 | KP683093 | KP852490 | gloeosporioides |
| C. chlorophyti# | IMI 103806* | GU227894 | GU228286 | GU228384 | GU227992 | GU228188 | singleton |
| C. chiangraiense | MFLUCC 14-0119* | MF448522 | MH376383 | MH351275 | gigapsorum | ||
| C. chrysanthemi# | IMI 364540* | JQ948273 | JQ948603 | JQ948934 | JQ949594 | JQ949924 | acutatum |
| C. chrysophillum | URM7368* | KX094252 | KX094183 | KX094083 | KX093982 | KX094285 | gloeosporioides |
| C. ciggaro# | ICMP 18539* | JX010230 | JX009966 | JX009800 | JX009523 | JX010434 | gloeosporioides |
| C. circinans | CBS 221.81 | GU227855 | GU228247 | GU228345 | GU227953 | GU228149 | dematium |
| C. citri | CBS 134233* | KC293581 | KC293741 | KY856138 | KY855973 | KC293661 | acutatum |
| C. citricola# | CBS 134228* | KC293576 | KC293736 | KC293792 | KC293616 | KC293656 | boninense |
| C. clidemiae | ICMP 18658* | JX010265 | JX009989 | JX009877 | JX009537 | JX010438 | gloeosporioides |
| C. cliviicola# | CBS 125375* | MG600733 | MG600795 | MG600850 | MG600939 | MG601000 | orchidearum |
| C. cobbittiense | BRIP 66219a* | MH087016 | MH094133 | MH094135 | MH094134 | MH094137 | gloeosporioides |
| C. coccodes# | CBS 369.75* | HM171679 | HM171673 | JQ005796 | HM171667 | JQ005859 | singleton |
| C. coelogynes | CBS 132504* | MG600713 | MG600776 | MG600836 | MG600920 | MG600980 | dracaenophilum |
| C. colombiense | CBS 129818* | JQ005174 | JQ005261 | JQ005348 | JQ005522 | JQ005608 | boninense |
| C. condaoense | CBS 134299* | MH229914 | MH229920 | MH229926 | MH229923 | boninense | |
| C. conoides# | CAUG17* | KP890168 | KP890162 | KP890156 | KP890144 | KP890174 | gloeosporioides |
| C. constrictum | CBS 128504* | JQ005238 | JQ005325 | JQ005412 | JQ005586 | JQ005672 | boninense |
| C. cordylinicola# | MFU 09-0551* | JX010226 | JX009975 | JX009864 | HM470234 | JX010440 | gloeosporioides |
| C. cosmi | CBS 853.73* | JQ948274 | JQ948604 | JQ948935 | JQ949595 | JQ949925 | acutatum |
| C. costaricense | CBS 330.75* | JQ948180 | JQ948510 | JQ948841 | JQ949501 | JQ949831 | acutatum |
| C. curcumae# | IMI 288937* | GU227893 | GU228285 | GU228383 | GU227991 | GU228187 | truncatum |
| C. cuscutae | IMI 304802* | JQ948195 | JQ948525 | JQ948856 | JQ949516 | JQ949846 | acutatum |
| C. cymbidiicola# | IMI 347923* | JQ005166 | JQ005253 | JQ005340 | JQ005514 | JQ005600 | boninense |
| C. dacrycarpi | CBS 130241* | JQ005236 | JQ005323 | JQ005410 | JQ005584 | JQ005670 | boninense |
| C. dematium# | CBS 125.25* | GU227819 | GU228211 | GU228309 | GU227917 | GU228113 | dematium |
| C. destructivum# | CBS 136228* | KM105207 | KM105561 | KM105277 | KM105417 | KM105487 | destructivum |
| C. doitungense | MFLUCC 14-0128* | MF448524 | MH049480 | MH376385 | MH351277 | boninense | |
| C. dracaenophilum# | CBS 118199* | JX519222 | JX546707 | JX519230 | JX519238 | JX519247 | dracaenophilum |
| C. duyunensis | CGMCC 3.15105* | JX625160 | KC843515 | KC843530 | JX625187 | caudatum | |
| C. echinochloae | MAFF 511473* | AB439811 | graminicola | ||||
| C. eleusines | MAFF 511155* | JX519218 | JX519226 | JX519234 | JX519243 | graminicola | |
| C. endophytica# | MFLUCC 13–0418* | KC633854 | KC832854 | KF306258 | gloeosporioides | ||
| C. endophytum | CGMCC 3.15108 * | JX625177 | KC843521 | KC843533 | JX625206 | graminicola | |
| C. eremochloae# | CBS 129661* | JX519220 | JX519228 | JX519236 | JX519245 | graminicola | |
| C. eriobotryae# | GLMC 1935* | MF772487 | MF795423 | MN191653 | MN191648 | MF795428 | acutatum |
| C. eryngiicola | MFLUCC 17-0318* | KY792726 | KY792723 | KY792720 | KY792717 | KY792729 | dematium |
| C. excelsum-altitudinum | CGMCC 3.15130* | HM751815 | KC843502 | KC843548 | JX625211 | dracaenophilum | |
| C. euphorbiae | CBS 134725* | KF777146 | KF777131 | KF777128 | KF777125 | KF777247 | singleton |
| C. falcatum# | CBS 147945* | JQ005772 | JQ005793 | JQ005835 | JQ005856 | graminicola | |
| C. feijoicola | CBS 144633 * | MK876413 | MK876475 | MK876466 | MK876507 | boninense | |
| C. fioriniae# | CBS 128517* | JQ948292 | JQ948622 | JQ948953 | JQ949613 | JQ949943 | acutatum |
| C. fructi | CBS 346.37* | GU227844 | GU228236 | GU228334 | GU227942 | GU228138 | dematium |
| C. fructicola# | ICMP 18581* | JX010165 | JX010033 | JX009866 | FJ907426 | JX010405 | gloeosporioides |
| C. fructivorum# | CBS 133125* | JX145145 | JX145196 | gloeosporioides | |||
| C. fuscum# | CBS 133701* | KM105174 | KM105524 | KM105244 | KM105384 | KM105454 | destructivum |
| C. fusiforme | MFLU 13-0291* | KT290266 | KT290255 | KT290253 | KT290251 | KT290256 | truncatum |
| C. gigasporum# | CBS 133266* | KF687715 | KF687822 | KF687761 | KF687866 | gigasporum | |
| C. gloeosporioides# | CBS 112999* | JQ005152 | JQ005239 | JQ005326 | JQ005500 | JQ005587 | gloeosporioides |
| C. godetiae# | CBS 133.44* | JQ948402 | JQ948733 | JQ949063 | JQ949723 | JQ950053 | acutatum |
| C. graminicola# | M 1.001* | JQ005767 | JQ005788 | JQ005830 | JQ005851 | graminicola | |
| C. grevilleae | CBS 132879* | KC297078 | KC297010 | KC296987 | KC296941 | KC297102 | gloeosporioides |
| C. grossum# | CAUG7* | KP890165 | KP890159 | KP890153 | KP890141 | KP890171 | gloeosporioides |
| C. guajavae | IMI 350839* | JQ948270 | JQ948600 | JQ948931 | JQ949591 | JQ949921 | acutatum |
| C. guizhouensis | CGMCC 3.15112* | JX625158 | KC843507 | KC843536 | JX625185 | spaethianum | |
| C. hanaui | MAFF 305404* | JX519217 | JX519225 | JX519242 | graminicola | ||
| C. hebeiense# | MFLUCC13–0726* | KF156863 | KF377495 | KF289008 | KF377532 | KF288975 | gloeosporioides |
| C. hedericola | MFLU 15-0689* | MN631384 | MN635794 | MN635795 | gloeosporioides | ||
| C. helleniense# | CBS 142418* | KY856446 | KY856270 | KY856186 | KY856019 | KY856528 | gloeosporioides |
| C. henanense# | CGMCC 3.17354* | KJ955109 | KJ954810 | KM023257 | KJ955257 | gloeosporioides | |
| C. hemerocallidis | CDLG5 | JQ400005 | JQ400012 | Q399998 | JQ399991 | JQ400019 | dematium |
| C. higginsianum# | IMI 349061* | KM105184 | KM105535 | KM105254 | KM105394 | KM105464 | destructivum |
| C. hippeastri | CBS 125376* | JQ005231 | JQ005318 | JQ005405 | JQ005579 | JQ005665 | boninense |
| C. horii# | NBRC 7478* | GQ329690 | GQ329681 | JX009752 | JX009438 | JX010450 | gloeosporioides |
| C. hystricis# | CBS 142411* | KY856450 | KY856274 | KY856190 | KY856023 | KY856532 | gloeosporioides |
| C. hsienjenchang | MAFF 243051 | AB738855 | AB738846 | AB738845 | singleton | ||
| C. incanum | ATCC 64682* | KC110789 | KC110807 | KC110825 | KC110816 | spaethianum | |
| C. indonesiense | CBS 127551* | JQ948288 | JQ948618 | JQ948949 | JQ949609 | JQ949939 | acutatum |
| C. insertae | MFLU 15-1895* | KX618686 | KX618684 | KX618683 | KX618682 | KX618685 | dematium |
| C. jacksonii | MAFF 305460* | JX519216 | JX519224 | JX519233 | JX519241 | graminicola | |
| C. jasminigenum | CGMCC LLTX-01* | HM131513 | HM131499 | HM131508 | HM153770 | truncatum | |
| C. javanense# | CBS 144963a* | MH846576 | MH846572 | MH846573 | MH846575 | MH846574 | acutatum |
| C. jiangxiense | CGMCC 3.17363* | KJ955201 | KJ954902 | KJ954471 | KJ955348 | gloeosporioides | |
| C. jinshuiense# | CGMCC 3.18903* | MG748077 | MG747995 | MG747913 | MG747767 | MG748157 | dematium |
| C. jishouense | GMBC0209* | MH482929 | MH681658 | MH708135 | MH727473 | gigasporum | |
| C. johnstonii | CBS 128532* | JQ948444 | JQ948775 | JQ949105 | JQ949765 | JQ950095 | acutatum |
| C. kahawae# | IMI 319418* | JX010231 | JX010012 | JX009813 | JX009452 | JX010444 | gloeosporioides |
| C. kakivorum# | KCTC 46679* | LC324781 | LC324787 | LC324783 | LC324785 | LC324791 | dematium |
| C. karstii# | CORCG6* | HM585409 | HM585391 | HM582023 | HM581995 | HM585428 | boninense |
| C. kinghornii | CBS 198.35* | JQ948454 | JQ948785 | JQ949115 | JQ949775 | JQ950105 | acutatum |
| C. laticiphilum# | CBS 112989* | JQ948289 | JQ948619 | JQ948950 | JQ949610 | JQ949940 | acutatum |
| C. lauri | MFLUCC 17-0205* | KY514347 | KY514344 | KY514341 | KY514338 | KY514350 | acutatum |
| C. ledebouriae | CBS 141284* | KX228254 | KX228357 | singleton | |||
| C. lentis# | CBS 127604* | JQ005766 | KM105597 | JQ005787 | JQ005829 | JQ005850 | destructivum |
| C. liaoningense# | CGMCC 3.17616* | KP890104 | KP890135 | KP890127 | KP890097 | KP890111 | magnum |
| C. lilii | CBS 109214/BBA 62147* | GU227810 | GU228202 | GU228300 | GU227908 | GU228104 | spaethianum |
| C. lini | CBS 172.51* | JQ005765 | KM105581 | JQ005786 | JQ005828 | JQ005849 | destructivum |
| C. limetticola | CBS 114.14* | JQ948193 | JQ948523 | JQ948854 | JQ949514 | JQ949844 | acutatum |
| C. limonicola# | CBS 142410* | KY856472 | KY856296 | KY856213 | KY856045 | KY856554 | boninense |
| C. lindemuthianum# | CBS 144.31* | JQ005779 | JX546712 | JQ005800 | JQ005842 | JQ005863 | orbiculare |
| C. lineola# | CBS 125337* | GU227829 | GU228221 | GU228319 | GU227927 | GU228123 | dematium |
| C. liriopes# | CBS 119444* | GU227804 | GU228196 | GU228294 | GU227902 | GU228098 | spaethianum |
| C. lobatum | IMI 79736* | MG600768 | MG600828 | MG600874 | MG600972 | MG601035 | magnum |
| C. lupini# | CBS 109225* | JQ948155 | JQ948485 | JQ948816 | JQ949476 | JQ949806 | acutatum |
| C. magnisporum | CBS 398.84* | KF687718 | KF687842 | KF687782 | KF687803 | KF687882 | gigasporum |
| C. makassarense# | CBS 143664a* | MH728812 | MH728820 | MH805850 | MH781480 | MH846563 | gloeosporioides |
| C. magnum | CBS 519.97* | MG600769 | MG600829 | MG600875 | MG600973 | MG601036 | magnum |
| C. malvarum | CBS 521.97* | KF178480 | KF178504 | KF178529 | KF178577 | KF178601 | orbiculare |
| C. melonis# | CBS 159.84* | JQ948194 | JQ948524 | JQ948855 | JQ949515 | JQ949845 | acutatum |
| C. menispermi | MFLU 14-0625* | KU242357 | KU242356 | KU242355 | KU242353 | KU242354 | dematium |
| C. metake | MAFF 244029 | AB738859 | singleton | ||||
| C. merremiae | CBS 124955* | MG600765 | MG600825 | MG600872 | MG600969 | MG601032 | magnum |
| C. miscanthi | MAFF 510857* | JX519221 | JX519229 | JX519237 | JX519246 | graminicola | |
| C. musae# | CBS 116870* | HQ596292 | HQ596299 | JX009896 | HQ596284 | HQ596280 | gloeosporioides |
| C. musicola# | CBS 132885* | MG600736 | MG600798 | MG600853 | MG600942 | MG601003 | orchidearum |
| C. navitas | CBS 125086* | JQ005769 | JQ005790 | JQ005832 | JQ005853 | graminicola | |
| C. neosansevieriae | CPC 25127* | KR476747 | KR476791 | KR476790 | KR476797 | singleton | |
| C. nicholsonii | MAFF 511115* | JQ005770 | JQ005791 | JQ005833 | JQ005854 | graminicola | |
| C. nigrum# | CBS 169.49* | JX546838 | JX546742 | JX546693 | JX546646 | JX546885 | singleton |
| C. novae-zelandiae# | CBS 128505* | JQ005228 | JQ005315 | JQ005402 | JQ005576 | JQ005662 | boninense |
| C. nupharicola# | CBS 470.96* | JX010187 | JX009972 | JX009835 | JX009437 | JX010398 | gloeosporioides |
| C. nymphaeae# | CBS 515.78* | JQ948197 | JQ948527 | JQ948858 | JQ949518 | JQ949848 | acutatum |
| C. ocimi | CBS 298.94* | KM105222 | KM105577 | KM105292 | KM105432 | KM105502 | destructivum |
| C. ochracea | CGMCC 3.15104* | JX625156 | KC843513 | KC843527 | JX625183 | caudatum | |
| C. okinawense# | MAFF 240517* | MG600767 | MG600827 | MG600971 | MG601034 | magnum | |
| C. oncidii | CBS 129828* | JQ005169 | JQ005256 | JQ005343 | JQ005517 | JQ005603 | boninense |
| C. orbiculare# | CBS 570.97* | KF178466 | KF178490 | KF178515 | KF178563 | KF178587 | orbiculare |
| C. orchidearum# | CBS 135131* | MG600738 | MG600800 | MG600855 | MG600944 | MG601005 | orchidearum |
| C. orchidophilum# | CBS 632.80* | JQ948151 | JQ948481 | JQ948812 | JQ949472 | JQ949802 | singleton |
| C. orchidis | MFLUCC 17-1302* | MK502144 | MK496857 | MK496855 | MK496853 | MK496859 | dematium |
| C. panacicola | C08087* | GU935869 | GU935849 | GU944758 | GU935889 | destructivum | |
| C. pandanicola | MFLUCC 17-0571* | MG646967 | MG646934 | MG646931 | MG646938 | MG646926 | gloeosporioides |
| C. panamense | CBS 125386* | MG600766 | MG600826 | MG600873 | MG600970 | MG601033 | magnum |
| C. paranaense# | CBS 134729* | KC204992 | KC205026 | KC205043 | KC205077 | KC205060 | acutatum |
| C. parallelophorum | MFLUCC 14-0083* | MF448525 | MK165695 | MH351280 | singleton | ||
| C. parsonsiae | CBS 128525* | JQ005233 | JQ005320 | JQ005407 | JQ005581 | JQ005667 | boninense |
| C. parthenocissicola | MFLUCC 17-1098* | MK629452 | MK639362 | MK639356 | MK639358 | MK639360 | dematium |
| C. paspali | MAFF 305403* | JX519219 | JX519227 | JX519235 | JX519244 | graminicola | |
| C. paxtonii | IMI 165753* | JQ948285 | JQ948615 | JQ948946 | JQ949606 | JQ949936 | acutatum |
| C. petchii | CBS 378.94* | JQ005223 | JQ005310 | JQ005397 | JQ005571 | JQ005657 | boninense |
| C. perseae# | CBS 141365* | KX620308 | KX620242 | KX620145 | KX620341 | gloeosporioides | |
| C. phaseolorum# | CBS 157.36 | GU227896 | GU228288 | GU228386 | GU227994 | GU228190 | dematium |
| C. phormii | CBS 118194* | JQ948446 | JQ948777 | JQ949107 | JQ949767 | JQ950097 | acutatum |
| C. phyllanthi# | CBS 175.67* | JQ005221 | JQ005308 | JQ005395 | JQ005569 | JQ005655 | boninense |
| C. piperis# | IMI 71397* | MG600760 | MG600820 | MG600867 | MG600964 | MG601027 | orchidearum |
| C. pisicola | CBS 724.97* | KM105172 | KM105522 | KM105242 | KM105382 | KM105452 | destructivum |
| C. plurivorum# | CBS 125474* | MG600718 | MG600781 | MG600841 | MG600925 | MG600985 | orchidearum |
| C. pseudoacutatum# | CBS 436.77* | JQ948480 | JQ948811 | JQ949141 | JQ949801 | JQ950131 | singleton |
| C. pseudomajus | CBS 571.88* | KF687722 | KF687826 | KF687779 | KF687801 | KF687883 | gigasporum |
| C. pseudotheobromicola# | MFLUCC 18–1602* | MH817395 | MH853675 | MH853678 | MH853681 | MH853684 | gloeosporioides |
| C. psidii | CBS 145.29* | JX010219 | JX009967 | JX009901 | JX009515 | JX010443 | gloeosporioides |
| C. proteae | CBS 132882* | KC297079 | KC297009 | KC296986 | KC296940 | KC297101 | gloeosporioides |
| C. pyricola | CBS 128531* | JQ948445 | JQ948776 | JQ949106 | JQ949766 | JQ950096 | acutatum |
| C. pyrifoliae# | CGMCC 3.18902* | MG748078 | MG747996 | MG747914 | MG747768 | MG748158 | singleton |
| C. queenslandicum# | ICMP 1778* | JX010276 | JX009934 | JX009899 | JX009447 | JX010414 | gloeosporioides |
| C. radicis | CBS 529.93* | KF687719 | KF687825 | KF687762 | KF687785 | KF687869 | gigasporum |
| C. rhexiae | CBS 133134* | JX145128 | JX145179 | gloeosporioides | |||
| C. rhombiforme# | CBS 129953* | JQ948457 | JQ948788 | JQ949118 | JQ949778 | JQ950108 | acutatum |
| C. riograndense | ICMP 20083* | KM655299 | KM655298 | KM655297 | KM655295 | KM655300 | spaethianum |
| C. roseum | CBS 145754 * | MK903611 | MK903603 | MK903604 | MK903607 | acutatum | |
| C. rusci | CBS 119206* | GU227818 | GU228210 | GU228308 | GU227916 | GU228112 | singleton |
| C. salicis# | CBS 607.94* | JQ948460 | JQ948791 | JQ949121 | JQ949781 | JQ950111 | acutatum |
| C. salsolae# | ICMP 19051* | JX010242 | JX009916 | JX009863 | JX009562 | JX010403 | gloeosporioides |
| C. sambucicola | MFLUCC 16–1388* | KY098781 | KY098780 | KY098779 | KY098778 | KY098782 | dematium |
| C. sansevieriae# | MAFF 239721/Sa-1-2 | LC179806 | LC180130 | LC180129 | LC180127 | LC180128 | singleton |
| C. scovillei# | CBS 126529* | JQ948267 | JQ948597 | JQ948928 | JQ949588 | JQ949918 | acutatum |
| C. sedi | MFLUCC14-1002* | KM974758 | KM974755 | KM974754 | KM974756 | KM974757 | dematium |
| C. serranegrense | COAD 2100a* | KY400111 | KY407894 | KY407892 | KY407896 | gigasporum | |
| C. shisoi# | JCM 31818* | MH660930 | MH660931 | MH660929 | MH660928 | MH660932 | destructivum |
| C. siamense# | MFU 090230* | FJ972613 | FJ972575 | JX009865 | FJ907423 | FJ907438 | gloeosporioides |
| C. sidae | CBS 504.97* | KF178472 | KF178497 | KF178521 | KF178569 | KF178593 | orbiculare |
| C. simmondsii# | CBS 122122* | JQ948276 | JQ948606 | JQ948937 | JQ949597 | JQ949927 | acutatum |
| C. sloanei | IMI 364297* | JQ948287 | JQ948617 | JQ948948 | JQ949608 | JQ949938 | acutatum |
| C. somersetense | CBS 131599* | JX076862 | caudatum | ||||
| C. sonchicola | MFLUCC 17-1299* | KY962757 | KY962754 | KY962751 | KY962748 | dematium | |
| C. sojae | ATCC 62257* | MG600749 | MG600810 | MG600860 | MG600954 | MG601016 | orchidearum |
| C. spaethianum# | CBS 167.49* | GU227807 | GU228199 | GU228297 | GU227905 | GU228101 | spaethianum |
| C. spinaciae# | CBS 128.57 | GU227847 | GU228239 | GU228337 | GU227945 | GU228141 | dematium |
| C. spinosum | CBS 515.97* | KF178474 | KF178498 | KF178523 | KF178571 | KF178595 | orbiculare |
| C. sublineola# | CBS 131301* | JQ005771 | JQ005792 | JQ005834 | JQ005855 | graminicola | |
| C. sydowii | CBS135819* | KY263783 | KY263785 | KY263787 | KY263791 | KY263793 | singleton |
| C. syzygiicola | MFLUCC 10-0624* | KF242094 | KF242156 | KF157801 | KF254880 | gloeosporioides | |
| C. tabacum | CPC 18945* | KM105204 | KM105557 | KM105274 | KM105414 | KM105484 | destructivum |
| C. tainanense | CBS 143666a* | MH728818 | MH728823 | MH805845 | MH781475 | MH846558 | gloeosporioides |
| C. tanaceti# | CBS 132693* | JX218228 | JX218243 | JX259268 | JX218238 | JX218233 | destructivum |
| C. tamarilloi# | CBS 129814* | JQ948184 | JQ948514 | JQ948845 | JQ949505 | JQ949835 | acutatum |
| C. tebeesti | CBS 522.97* | KF178473 | KF178505 | KF178522 | KF178570 | KF178594 | orbiculare |
| C. theobromicola# | ICMP 18649* | JX010294 | JX010006 | JX009869 | JX009444 | JX010447 | gloeosporioides |
| C. temperatum | CBS 133122* | JX145159 | JX145211 | gloeosporioides | |||
| C. ti# | ICMP 4832* | JX010269 | JX009952 | JX009898 | JX009520 | JX010442 | gloeosporioides |
| C. tofieldiae | CBS 495.85* | GU227801 | GU228193 | GU228291 | GU227899 | GU228095 | spaethianum |
| C. tongrenense | GZU_TRJ1-37/GMBC0209* | MH482933 | MH705332 | MH717074 | MH729805 | dracaenophilum | |
| C. torulosum | CBS 128544* | JQ005164 | JQ005251 | JQ005338 | JQ005512 | JQ005598 | boninense |
| C. trichellum | CBS 217.64* | GU227812 | GU228204 | GU228302 | GU227910 | GU228106 | singleton |
| C. trifolii# | CBS 158.83* | KF178478 | KF178502 | KF178527 | KF178575 | KF178599 | orbiculare |
| C. tropicale# | CBS 124949* | JX010264 | JX010007 | JX009870 | JX009489 | JX010407 | gloeosporioides |
| C. tropicicola | BCC 38877* | JN050240 | JN050229 | JN050218 | JN050246 | dracaenophilum | |
| C. truncatum# | CBS 151.35* | GU227862 | GU228254 | GU228352 | GU227960 | GU228156 | truncatum |
| C. utrechtense | CBS 130243* | KM105201 | KM105554 | KM105271 | KM105411 | KM105481 | destructivum |
| C. verruculosum | IMI 45525* | GU227806 | GU228198 | GU228296 | GU227904 | GU228100 | spaethianum |
| C. vietnamense | LD16(L2)* | KF687721 | KF687832 | KF687769 | KF687792 | KF687877 | gigasporum |
| C. vignae | CBS 501.97* | KM105183 | KM105534 | KM105253 | KM105393 | KM105463 | destructivum |
| C. viniferum# | GZAAS5.08601* | JN412804 | JN412798 | JN412795 | JN412813 | gloeosporioides | |
| C. vittalense | CBS 181.82* | MG600734 | MG600796 | MG600851 | MG600940 | MG601001 | orchidearum |
| C. walleri | CBS 125472* | JQ948275 | JQ948605 | JQ948936 | JQ949596 | JQ949926 | acutatum |
| C. wanningense# | CGMCC 3.18936* | MG830462 | MG830318 | MG830302 | MG830270 | MG830286 | acutatum |
| C. watphraense | MFLUCC 14-0123* | MF448523 | MH049479 | MH376384 | MH351276 | boninense | |
| C. wuxiense# | CGMCC 3.17894* | KU251591 | KU252045 | KU251939 | KU251672 | KU252200 | gloeosporioides |
| C. xanthorrhoeae | BRIP 45094* | JX010261 | JX009927 | JX009823 | JX009478 | JX010448 | gloeosporioides |
| C. yulongense | CFCC 50818* | MH751507 | MK108986 | MH793605 | MH777394 | MK108987 | gloeosporioides |
| C. yunnanense | CBS 132135* | JX546804 | JX546706 | JX519231 | JX519239 | JX519248 | dracaenophilum |
| C. zoysiae | MAFF238573* | JX076871 | caudatum |
Fig. 2 Phylogram generated from MP analysis based on combined sequences of ITS, gapdh, chs, act and tub2 sequences of all species of Colletotrichum with molecular data. Related sequences were obtained from GenBank. Two hundred and fourty nine taxa are included in the analyses, which comprise 2296 characters including gaps, of which 868 characters are constant, 295 characters are parsimony-uninformative and 1133 characters parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of ten equally most parsimonious trees with a length of 10088 steps (CI = 0.283, RI=0.840, RC = 0.237, HI = 0.717) in the third tree. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Monilochaetes infuscans (CBS 869.96) and M. populi (CBS 139623). MP bootstrap support value ≥50% and BYPP ≥0.9 are shown respectively near the nodes. Ex-type strains are in bold.


No Comments