26 Oct Meliola
Meliola Fr., Syst. orb. veg. (Lundae) 1: 111 (1825)
Background
Meliola commonly known as “black mildews” or “dark mildews” is the largest genus of Meliolaceae (Hongsanan et al. 2015; Zeng et al. 2017). Fries (1825) established this genus, with the type species M. nidulans. Species in Meliola are mostly biotrophs or pathogens of living leaves and occasionally petioles, twigs, and branches (Hansford 1961; Hosagoudar 1994, 1996, 2008; Mibey and Hawksworth 1997; Old et al. 2003; Hosagoudar and Riju 2013). The phylogenetic placement of Meliola was established by using sequence data from fruiting bodies and placed in Sordariomycetes (Gregory and John 1999; Pinho et al. 2012, 2014, Hongsanan et al. 2015; Justavino et al. 2015). Meliola has been shown to be polyphyletic (Hyde et al. 2020b; Marasinghe et al. 2020; Zeng et al. 2020). There is little sequence data available in GenBank for clarifying relationships between species and establishiing host-specificity (Hongsanan et al. 2015; Zeng et al. 2017).
Classification – Ascomycota, Pezizomycotina, Sordariomycetes, Sordariomycetidae, Meliolales, Meliolaceae
Type species – Meliola nidulans (Schwein.) Cooke
Distribution – commonly found in tropical and subtropical regions (see Zeng et al. 2017)
Disease symptoms – Black mildews, forming black, radiate velvety colonies on the surface of plants.
Hosts – has a wide range of hosts (see Zeng et al. 2017)
Pathogen biology, disease cycle and epidemiology
For pathogen biology, disease cycle and epidemiology see Hongsanan et al. (2015).
Morphological based identification and diversity
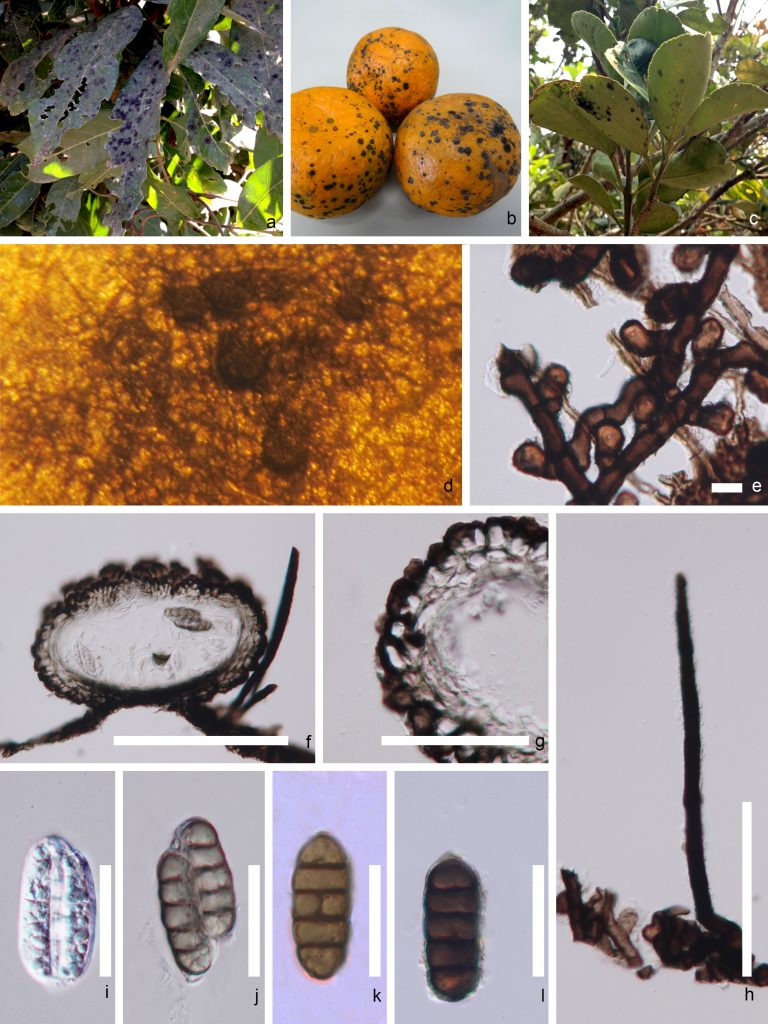
Species in Meliola are characterized by forming web-like colonies on the host surface, hyphal setae developed from superficial hyphae, with hyphopodia, 2–4-spored, unitunicate asci, and 3–4-septate pigmented ascospores (Pinho et al. 2012, 2014; Hongsanan et al. 2015, 2020; Justavino et al. 2015; Hyde et al. 2020; Fig. 1). Cannon and Kirk (2007) reported that the asexual morph of the genus develops from the hypha, form ampuliform hyphopodia or flask-shaped which are called “phialides” (Hongsananet et al. 2015). Conidiogenous cells formed from vegetative hyphae and small, hyaline, unicellular conidia (Cannon and Kirk 2007; Hongsanan et al. 2015). Currently, Meliola comprises over 1700 species (Zeng et al. 2017), which have mostly been introduced by host association, followed by morphology, and disease distribution (Mibey and Hawksworth 1997). Thus, species identification is almost impossible without a host name. However, the same species can be found in different hosts, but it is not clear if this is widespread (Hongsanan et al. 2015). Therefore, testing of host-specificity in Meliola is needed to establish accurate species determination.
Fig. 1 Morphology of Meliola species a Meliola thailandicum on Dimocarpus longan. b Meliola sp. on Citrus reticulata. c Meliola sp. on Citrus maxima. d Colony on the host surface. e Hyphopodia on mycelium. f Section through ascoma. g Peridium. h Setae. i Young ascus. j Mature ascus. k, l Ascospores.Scale bars: f=50 μm, g,i-l=30 μm, h=10 μm and e=5 μm.
Molecular based identification and diversity
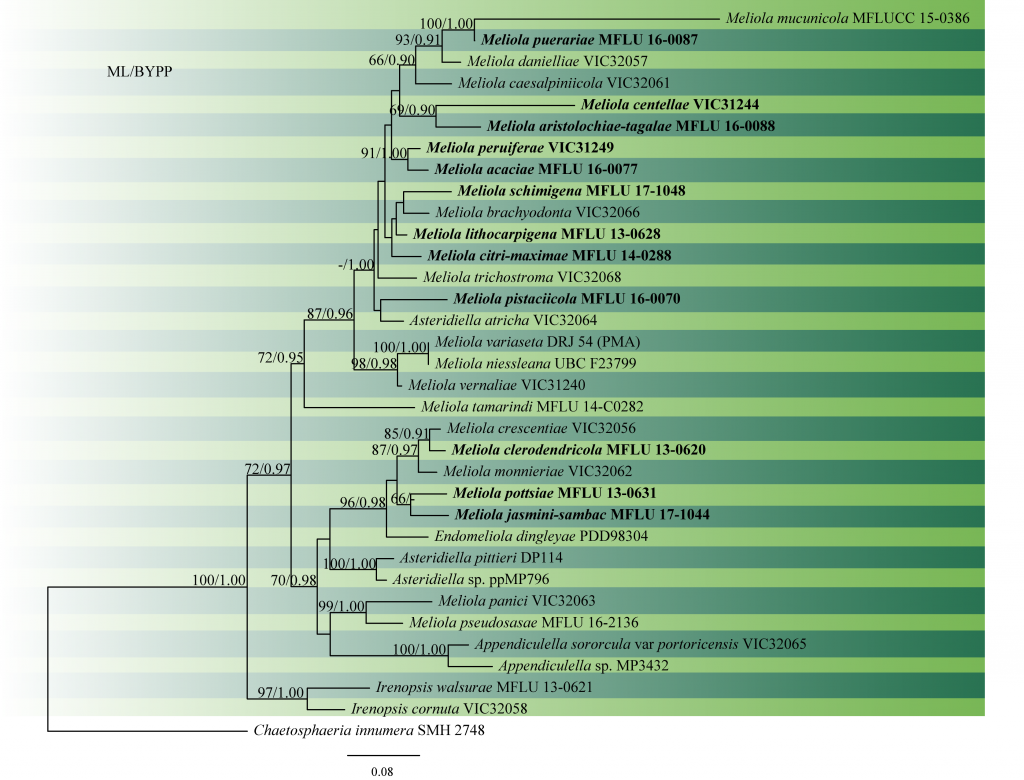
Sequence data of species in Meliola are from direct sequencing of fruiting bodies and fresh mycelium (Pinho et al. 2012, 2014; Hongsanan et al. 2015; Justavino et al. 2015; Hyde et al. 2016, 2020b). LSU and ITS sequence data placed Meliola in Sordariomycetes (Hongsanan et al. 2015, 2020; Maharachchikumbura et al. 2015, 2016; Hyde et al. 2016, 2020). By adding more sequence data, Meliola was shown to be polyphyletic (Marasinghe et al. 2020; Zeng et al. 2020). A phylogenetic tree for Meliola species is presented in Fig. 2.
Recommended genetic markers (genus level) – LSU, SSU of nrDNA
Recommended genetic marker (species level) – ITS
Accepted number of species – There are 3064 epithets listed in Index Fungorum (2020), however only 25 species have DNA molecular data (Zeng et al. 2017, Table 1).
References – Cannon and Kirk 2007 (morphology); Pinho et al. 2012, 2014, Hongsanan et al. 2015, 2020, Justavino et al. 2015, Zeng et al. 2020 (morphology and phylogeny).
Table 1 DNA barcodes available for Meliola. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Species confirmed with pathogenicity studies are marked with #.
| Species | Culture/ Specimen number | LSU | ITS |
| MN747479 | MN747479 | ||
| Meliola aristolochiae-tagalae | MFLU 16-0088* | MN747473 | – |
| M. brachyodonta | VIC32066 | KC618644 | – |
| M. caesalpiniicola | VIC32061 | KC618641 | – |
| M. centellae | VIC31244* | NG042650 | NR137799 |
| M. citri-maximae | MFLU 14-0288* | KX458474 | – |
| M. clerodendricola | MFLU 13-0620* | KT021647 | – |
| M. crescentiae | VIC32056 | KC618649 | – |
| M. danielliae | VIC32057 | KC618648 | – |
| M. jasmini-sambac | MFLU 17-1044* | MN747482 | MN747482 |
| M. lithocarpigena | MFLU 13-0628* | MN747474 | MN747474 |
| M. monnieriae | VIC32062 | KC618647 | – |
| M. mucunicola | MFLU 15-0386 | KT157533 | KT157534 |
| M. niessleana | UBC F23799 | KC833049 | – |
| M. panici | VIC32063 | KC618651 | – |
| M. peruiferae | VIC 31249* | NG 060294 | – |
| M. pistaciicola | MFLU 16-0070* | MN747478 | MN747478 |
| M. pottsiae | MFLU 13-0631* | MN747475 | MN747475 |
| M. puerariae | MFLU 16-0087* | MN747472 | – |
| M. pseudosasae | MFLU 16-2136 | KX845434 | – |
| M. schimigena | MFLU 17-1048* | MN747484 | MN747484 |
| M. tamarindi | MFLU 14-C0282 | KP744489 | – |
| M. trichostroma | VIC32068 | KC618643 | – |
| M. variaseta | DRJ 54 (PMA) | EF094840 | – |
| M. vernaliae | VIC 31240 | JX096808 | – |
Fig. 2 Phylogram generated from RAxML analysis based on combined ITS and LSU sequence data of Meliola species. Related sequences were obtained from GenBank. Thirty-five strains are included in the analyses, which comprised 1655 characters including gaps. The tree was rooted with Chaetosphaeria innumera (SMH 2748). Tree topology of the ML analysis was similar to the Bayesian analysis. ML bootstrap values ≥50% and BYPP ≥0.90 are shown respectively near the nodes.

No Comments