27 Oct Pythium
Pythium Pringsh., Jb. wiss. Bot. 1: 304 (1858)
Background
Pythium is the largest and most comprehensively studied genus in Pythiaceae sensu lato, order Peronosporales sensu lato, class Peronosporomycetes, phylum Oomycota, and kingdom Straminipila (Beakes et al. 2014). Pringsheim (1858) described the genus. However, the initial classification of Pythium has changed many times based on several studies using morphological characteristics (Uzuhashi et al. 2010). Pythium comprises of more than 230 extant species (Hyde et al. 2014), however, identification of species has always been problematic due to limited morphological characters, difficulty in isolating some taxa and lack of molecular data for certain species (Lévesque and de Cock 2004).
Classification – Oomycota, Pythiales, Pythiaceae
Type species – Pythium monospermum Pringsh. (Pringsheim 1858)
Distribution – worldwide
Disease symptoms – generally cause rot of fruit, roots and stem including pre- or post-emergence damping-off of seeds and seedlings.
Pythium causes crown and root rot in mature plants, where plants suddenly wilt during warm and sunny weather and when plants have their first heavy fruit load. Often, upper leaves of infected plants wilt in the day and recover overnight. However, plants eventually die (Craft and Nelson 1996; Postma et al. 2000). The first symptoms of Pythium root infections include stunting. In the root system, initial symptoms are brown to dark-brown lesions on root tips and feeder roots. As the disease progresses, symptoms are soft, brown, stubby roots and lack of feeder roots. In larger roots, the outer root tissue or cortex peels away, leaving the string-like vascular bundles underneath (Postma et al. 2000; Moorman et al. 2002; Al-Mahmooli et al. 2015). Pythium rot also occurs in the crown at the stem base. In cucumber, diseased crowns turn orange-brown, often with a soft rot at the base, while in strawberry seedling roots have dark brown, water-soaked rot and rotten crowns (Columbia and English 1988; Ishiguro et al. 2014). Several species of Pythium cause blight of turfgrass, which initially appears as “greasy” water-soaked areas, but later turn brown and grey (Vencelli and Powell 2008).
Several Pythium species are capable of causing fruit rot in numerous crops (Martin and Loper 1999). Pythium fruit rot is commonly known as a cottony leak or watery rot and occurs during wet weather or in poorly drained areas of fields (Ho and Abd-Elsalam 2020; Sharma et al. 2020a). Initial symptoms of the fruit rot are brownish, water-soaked lesions that quickly become large, watery, soft and rotten. The rot generally begins on the parts of fruit in contact with the soil. In cucumber, a brown to dark green blister can be seen on fruit before they become watery and rot. Later, white cottony mycelium can be seen on rotten tissues, especially during humid weather. Pythium fruit rot is most severe in poorly-drained fields during wet weather. The disease can render fruit unmarketable (Ho 2009; Sharma et al. 2020a).
Pre-emergence damping-off causes seeds and young seedlings to rot before they emerge from the growing medium in greenhouses, while post-emergence damping-off kills newly emerged seedlings. In the latter, the pathogen causes a water-soaked, soft brown lesion at the stem base, near the soil line, that pinches off the stem causing the seedling to topple over and die (Weiland et al. 2012).
Hosts– Pythium has a wide range of hosts including species of Cucurbitaceae and Poaceae, Ananas comosus, Arachis hypogaea, Brassica sp., Carica papaya, Beta vulgaris, Daucus carota subsp. sativus, Dendrobium sp, Solanum sp. and Zingiber officinale. Some species are pathogens of algae, fungi, other oomycetes, nematodes, insects, animals and humans (Van der Plaäts-Niterink1981; Czeczuga et al. 2005; Kawamura et al. 2005; Hwang et al. 2009; Li et al. 2010; Weiland et al. 2012; Ho 2013; Hyde et al 2014). Several species inhabit different soils in cultivated and uncultivated fields including forest (Uzuhashi et al 2010). Pythium arrhenomanes, P. dissotocum, P. elongatum, P. myriotylum, and P. spinosum are important pathogens of rice seedlings (Hendrix and Campbell 1973; Hsieh 1978; Ventura et al. 1981; Chun and Schneider 1998; Eberle et al. 2007; Kreye et al. 2009; Oliva et al. 2010; Banaay et al. 2012; Van Buyten and Höfte 2013). Pythium insidiosum causes pythiosis in mammals including humans (Van der Plaats-Niterink 1981; de Cock et al. 1987). Some species target below-ground plant parts and some species can cause fruit rot, however, some Pythium species can also benefit plants as endophytes by acting as biocontrol agents (Benhamou et al. 1997) and by stimulating plant growth (Martin and Looper 1999; Mazzola et al. 2002).
Pathogen biology, disease cycle and epidemiology
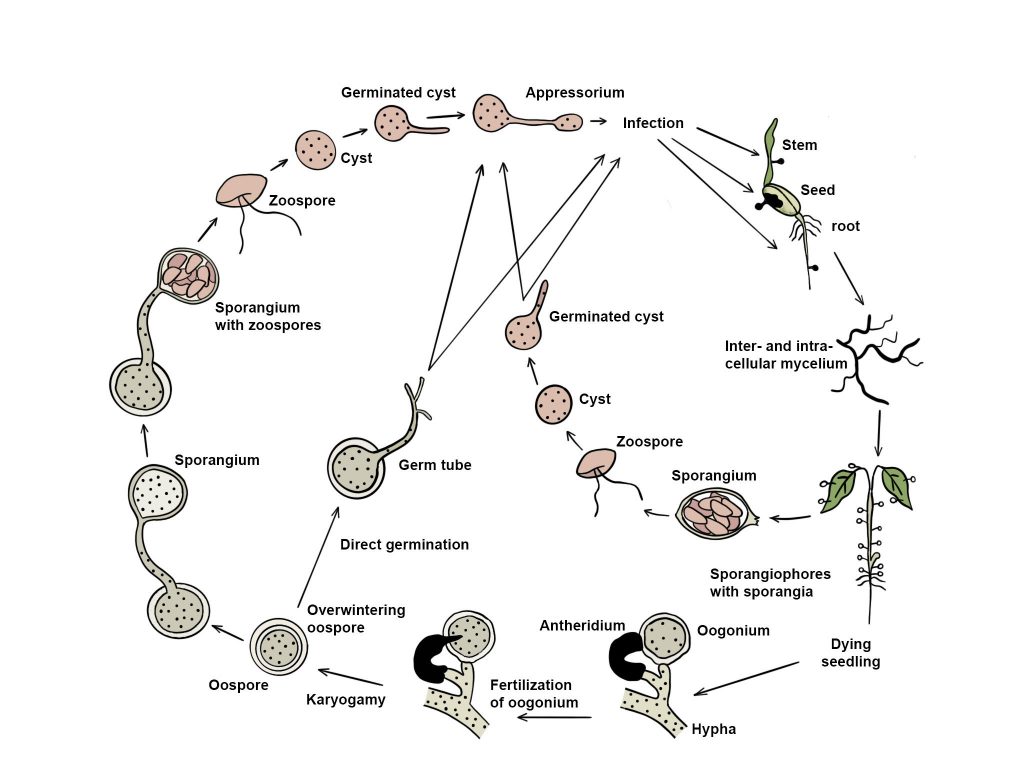
Pythium species grow and colonize a plant by producing hyphae which extract nutrients from the host. Once the hyphae from opposite mating types meet, they produce thick-walled oospores which serve as overwintering structures. Upon germination, an oospore may produce more hyphae, or develop a zoosporangium, which produces motile zoospores that swim to and infect plants. Zoosporangia can also germinate and directly infect plants (Ho 2009; van West et al. 2003).
Fig. 1 Disease cycle of a Pythium species (redrawn from van West et al. 2003)
Morphological based identification and diversity
Pythium has hyaline hyphae which are coenocytic without cross septa (van der Plaäts-Niterink 1981). Filamentous and globose sporangia are present, and zoospores develop in a vesicle, which is formed at the tip of a discharge tube from a sporangium. After fertilization with paragynous or hypogynous antheridia, oospores are formed in smooth or ornamented oogonia. The oospore can fill the whole organism or can have space between the walls of the oogonia and oospore. The process of zoospore formation within a vesicle is a characteristic feature of the genus, which distinguishes it from morphologically similar genera such as Phytophthora and Halophytophthora. However, the formation of zoospores is similar to Lagenidium, which features endobiotic and holocarpic features not observed in Pythium (Dick 2001). Species delimitation based on morphological characteristics such as shape and size of sporangia and oogonia is difficult as these characteristics are often shared among different species.
Molecular based identification and diversity
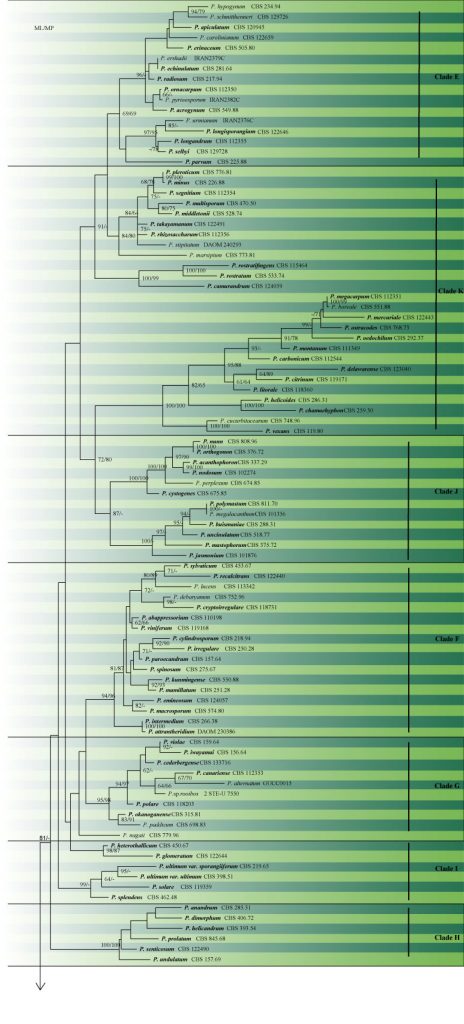
Lévesque and de Cock (2004) separated the genus into 11 clades (A-K) using phylogenies of ITS and 28S. Clade K, which includes P. vexans was transferred to a new genus Phytopythium with Phytopythium sindhum as type species (Bala et al. 2010), while the remaining clades can be divided into two groups: species with filamentous sporangia (clades A-D) and species with globose sporangia (clades E-J). Identification of Pythium isolates to species level is recommended based on cox1 and ITS gene regions. The use of ITS region alone cannot accurately identify all Pythium species. Several species are indistinguishable based on both ITS and cox1 sequences. Lévesque and de Cock (2004) provided the first extensive study of Pythium, accepting 116 species. Additional species have recently been described for example P. alternatum (Rahman et al. 2015), P. biforme, P. brachiatum, P. junctum, P. utonaiense (Uzuhashi et al. 2015), P. cedri (Chen et al. 2017a), P. heteroogonium, P. longipapillum, P. oryzicollum (Salmaninezhad et al. 2019). Currently, there are more than 130 accepted species in the genus (Arafa et al. 2020). The phylogenetic tree constructed is presented in Fig. 2 and the information of species are given in table 1.
Recommended genetic markers (generic level within Pythium sensu lato) – 18S (small subunit, SSU) and 28S (large subunit, LSU) nuclear rRNA genes
Recommended genetic markers (sub-generic, inter- and intra-specific level) – The internal transcribed spacers (ITS including ITS1, 5.8S rRNA, and ITS2), cytochrome c oxidase subunit 2 (cox2)
Accepted number of species – There are 330 epithets listed in Index Fungorum (2020), however only 157 species have DNA sequence data (Table 1).
References – van der Plaäts-Niterink 1981, Dick 2001 (morphology), Lévesque and de Cock 2004, Hyde et al. 2014, Arafa et al. 2020 (phylogeny and accepted species numbers).
Table 1 DNA barcodes available for Pythium. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains and voucher strains are in bold. Species confirmed with pathogenicity studies are marked with #. (VdPN) are strains used by Van der Plaäts-Niterink (1981) for descriptions.
| Species | Isolate | Host | SSU | ITS | LSU | cox2 | tub2 |
| Pythium abappressorium# | CBS 110198 | Triticum aestivum | HQ643408 | HQ643408 | HQ643408 | KJ595409 | KJ595533 |
| P. acanthicum# | CBS 377.34 | Solanum tuberosum | AY598617 | AY598617 | AY598617 | KJ595380 | KJ595504 |
| P. acanthophoron# | CBS 337.29 | Ananas sativus | AY598711 | AY598711 | AY598711 | KJ595376 | KJ595500 |
| P. acrogynum | CBS 549.88 | Soil (Spinacia oleracea) | N/A | AY598638 | AY598638 | AB362324 | KJ595458 |
| P. adhaerens | CBS 520.74 | Soil | AY598619 | AY598619 | AY598619 | KJ595386 | KJ595510 |
| P. afertile | LEV2066 | Turf grass | N/A | HQ643416 | HQ643416 | KJ595440 | KJ595563 |
| P. alternatum | GUCC0015 | Soil | N/A | AB998876 | N/A | – | N/A |
| P. amasculinum | CBS 552.88 | soil (vegetable garden) | AY598671 | AY598671 | AY598671 | KJ595390 | KJ595514 |
| P. anandrum# | CBS 285.31 | Rheum rhaponticum | AY598650 | AY598650 | AY598650 | AB362328 | KJ595450 |
| P. angustatum# | CBS 522.74 (VdPN) | Soil | AY598623 | AY598623 | AY598623 | KJ595387 | KJ595511 |
| P. aphanidermatum# | CBS 118.80 | Unknown | AY598622 | AY598622 | AY598622 | KJ595344 | KJ595472 |
| P. apiculatum | CBS 120945 | Soil (Vitis sp.) | HQ643443 | HQ643443 | HQ643443 | KJ595422 | KJ595547 |
| P. apleroticum | CBS 772.81 | Nymphyoidespeltata | AY598631 | AY598631 | AY598631 | KJ595400 | KJ595524 |
| P. aquatile# | CBS 215.80 | Soil | AY598632 | AY598632 | AY598632 | KJ595355 | KJ595481 |
| P. aristosporum | CBS 263.38 | Triticum aestivum | AY598627 | AY598627 | AY598627 | AB507410 | DQ071297 |
| P. arrhenomanes# | CBS 324.62 (VdPN) | Zea mays | AKXY02050628 | AY598628 | AY598628 | AKXY02053172 | KJ595451 |
| P. attrantheridium# | DAOM 230386 | Prunus serotina | HQ643476 | HQ643476 | HQ643476 | AB512889 | AB512822 |
| P. biforme | UZ00796 | Aquatic | N/A | KJ995584 | KJ995601 | N/A | N/A |
| P. boreale | CBS 551.88 | Soil | AY598662 | AY598662 | AY598662 | EF408876 | EF408882 |
| P. brachiatum | UZ00736 | Aquatic | N/A | KJ995581 | KJ995603 | N/A | N/A |
| P. buismaniae | CBS 288.31 | Linumusitatissimum | AY598659 | AY598659 | AY598659 | KJ595368 | KJ595493 |
| P. camurandrum | CBS 124059 | Hordeum vulgare | GQ244426 | GQ244426 | GQ244426 | KJ595433 | KJ595558 |
| P. canariense | CBS 112353 | Soil | HQ643482 | HQ643482 | HQ665069 | JX397983 | JX397969 |
| P. capillosum | CBS 222.94 | Soil | AY598635 | AY598635 | AY598635 | KJ595360 | KJ595485 |
| P. carbonicum# | CBS 112544 | Soil (spoil heap) | HQ643373 | HQ643373 | HQ643373 | AB690678 | KJ595464 |
| P. carolinianum# | CBS 122659 | Soil | N/A | HQ643484 | HQ665111 | KJ595427 | KJ595551 |
| P. catenulatum# | CBS 842.68 (VdPN) | Turf grass | AY598675 | AY598675 | AY598675 | KJ595404 | KJ595528 |
| P. caudatum# | CBS 584.85 | Xiphinemarivesi | HQ643136 | HQ643136 | HQ665277 | AF290309 | KJ595459 |
| P. cederbergense# | CBS 133716 | Aspalathuslinearis | N/A | JQ412768 | KJ716864 | JQ412805 | JQ412781 |
| P. cedri# | Chen 30 | roots of Cedrus | N/A | KX423751 | N/A | N/A | N/A |
| P. chamaehyphon# | CBS 259.30 | Carica papaya | AY598666 | AY598666 | AY598666 | AB257280 | KJ595448 |
| P. chondricola# | CBS 203.85 | Chondruscrispus | N/A | AY598620 | AY598620 | KJ595354 | KJ595480 |
| P. citrinum | CBS 119171 | Soil (Vitis sp.) | HQ643375 | HQ643375 | HQ643375 | AB690679 | KJ595465 |
| P. coloratum# | CBS 154.64 | Soil (tree nursery) | AY598633 | AY598633 | AY598633 | KJ595346 | KJ595474 |
| P. conidiophorum# | CBS 223.88 | Soil | AY598629 | AY598629 | AY598629 | KJ595361 | KJ595486 |
| P. contiguanum | CBS 221.94 | Soil (salt marsh) | HQ643514 | HQ643514 | HQ665162 | KJ595358 | KJ595483 |
| P. cryptoirregulare# | CBS 118731 | Euphorbia pulcherrima | HQ643515 | HQ643515 | HQ643515 | GU071763 | GU071888 |
| P. cucurbitacearum# | CBS 748.96 | Unknown | AY598667 | AY598667 | AY598667 | AB690680 | KJ595460 |
| P. cylindrosporum# | CBS 218.94 | Soil | AY598643 | AY598643 | AY598643 | GU071762 | GU071877 |
| P. cystogenes# | CBS 675.85 | Viciafaba | HQ643518 | HQ643518 | HQ643518 | KJ595396 | KJ595520 |
| P. debaryanum# | CBS 752.96 | Tulipa sp. | AY598704 | AY598704 | AY598704 | KJ595399 | KJ595523 |
| P. delawarense | CBS 123040 | Glycine max | KF853241 | EU339312 | KF853240 | KJ595430 | KJ595555 |
| P. deliense# | CBS 314.33 | Nicotiana tabacum | AY598674 | AY598674 | AY598674 | KJ595372 | KJ595497 |
| P. diclinum# | CBS 664.79 | Beta vulgaris | N/A | AY598690 | HQ665282 | KJ595394 | KJ595518 |
| P. dimorphum# | CBS 406.72 | Pinus taeda | AY598651 | AY598651 | AY598651 | AB362331 | KJ595454 |
| P. dissimile | CBS 155.64 | Pinus radiata | AY598681 | AY598681 | AY598681 | KJ595347 | KJ595475 |
| P. dissotocum# | CBS 166.68 (VdPN) | Triticum aestivum | AY598634 | AY598634 | AY598634 | KJ595351 | KJ595479 |
| P. echinulatum# | CBS 281.64 (VdPN) | Soil (forest nursery) | AY598639 | AY598639 | AY598639 | AB362327 | KJ595449 |
| P. emineosum | CBS 124057 | Juniperus communis | N/A | GQ244427 | GQ244427 | KJ595432 | KJ595557 |
| P. erinaceum | CBS 505.80 | Soil | N/A | AY598694 | HQ665243 | AB362326 | KJ595456 |
| P. ershadii | IRAN2379C | soil | N/A | KT894054 | N/A | – | N/A |
| P. flevoense | CBS 234.72 | Soil | AY598691 | AY598691 | AY598691 | KJ595363 | KJ595488 |
| P. folliculosum# | CBS 220.94 | Soil | AY598676 | AY598676 | HQ665160 | N/A | N/A |
| P. glomeratum# | CBS 122644 | Soil | N/A | HQ643542 | HQ665097 | KJ595424 | KJ595548 |
| P. graminicola# | CBS 327.62 | Saccharum officinarum | AY598625 | AY598625 | AY598625 | AF196593 | KJ595452 |
| P. grandisporangium# | CBS 286.79 | Decaying leaf (Zostera marina) | AY598692 | AY598692 | AY598692 | KJ595367 | KJ595492 |
| P. helicandrum | CBS 393.54 | Rumex acetosella | AY598653 | AY598653 | AY598653 | AB362329 | KJ595453 |
| P. helicoides# | CBS 286.31 | Phaseolus vulgaris | AY598665 | AY598665 | AY598665 | DQ071377 | AB511994 |
| P. heteroogonium | 079-1 – CBS 141232 | Soil | N/A | KX228103 | N/A | KX228131 | KX228117 |
| P. heterothallicum# | CBS 450.67 | Soil (Sambucus) | AY598654 | AY598654 | AY598654 | AB512919 | AB512850 |
| P. hydnosporum# | CBS 253.60 (VdPN) | Unknown | AY598672 | AY598672 | AY598672 | KJ595364 | KJ595489 |
| P. hypogynum# | CBS 234.94 | Soil | AY598693 | HQ643565 | HQ665171 | AB362325 | KJ595447 |
| P. inflatum# | CBS 168.68 (VdPN) | Saccharum officinarum | AY598626 | AY598626 | AY598626 | KJ595352 | N/A |
| P. insidiosum# | CBS 574.85 | Equus ferus | AF289981 | AY598637 | AY598637 | KJ595391 | KJ595515 |
| P. intermedium# | CBS 266.38 (VdPN) | Agrostis stolonifera | AY598647 | AY598647 | AY598647 | AB507410 | AB512836 |
| P. irregular# | CBS 250.28 | Phaseolus vulgaris | AY598702 | AY598702 | AY598702 | GU071760 | GU071886 |
| P. iwayamai# | CBS 156.64 (VdPN) | Soil (Pinus sp.) | AY598648 | AY598648 | AY598648 | JX397979 | JX397965 |
| P. jasmonium | CBS 101876 | Arabidopsis thaliana | HQ643778 | HQ643778 | HQ643778 | KJ595406 | KJ595530 |
| P. junctum | UZ00732 | Aquatic | N/A | KJ995576 | KJ995605 | N/A | N/A |
| P. kashmirense# | CBS 122908 | Soil | HQ643671 | HQ643671 | HQ643671 | KJ595429 | KJ595553 |
| P. kunmingense# | CBS 550.88 | Soil (Viciafaba) | AY598700 | AY598700 | HQ665259 | KJ595389 | KJ595513 |
| P. litorale# | CBS 118360 | Soil (Phragmites australis) | HQ643386 | HQ643386 | HQ643386 | KJ595418 | KJ595543 |
| P. longandrum# | CBS 112355 | Soil | HQ643679 | HQ643679 | HQ665071 | KJ595413 | KJ595538 |
| P. longipapillum# | NRh8* | soil | N/A | KX228105 | N/A | KX228130 | KX228114 |
| P. longisporangium# | CBS 122646 | Soil (Vitis sp.) | N/A | HQ643680 | HQ665099 | KJ595426 | KJ595550 |
| P. lucens# | CBS 113342 | Triticum | HQ643681 | HQ643681 | HQ643681 | KJ595415 | KJ595540 |
| P. lutarium# | CBS 222.88 | Soil | HQ643682 | HQ643682 | HQ665163 | KJ595359 | KJ595484 |
| P. lycopersici# | CBS 122909 | Soil (Lycopersicum esculentum) | N/A | HQ643683 | HQ665119 | KJ595343 | KJ595554 |
| P. macrosporum# | CBS 574.80 | Flower bulb | AY598646 | AY598646 | AY598646 | AB512916 | AB512842 |
| P. mamillatum# | CBS 251.28 (VdPN) | Beta vulgaris | AY598703 | AY598703 | HQ665173 | AB362325 | AB512844 |
| P. marinum# | CBS 750.96 | Soil | N/A | AY598689 | AY598689 | KJ595398 | KJ595522 |
| P. marsipium# | CBS 773.81 | Nymphyoides peltata | N/A | AY598699 | HQ665297 | KJ595401 | KJ595525 |
| P. mastophorum# | CBS 375.72 (VdPN) | Apiumgraveolens | AY598661 | AY598661 | AY598661 | KJ595378 | KJ595502 |
| P. megacarpum | CBS 112351 | Soil (Vitis sp.) | HQ643388 | HQ643388 | HQ643388 | AB690665 | KJ595536 |
| P. megalacanthum# | CBS 101356 | Chrysanthemum | N/A | HQ643693 | KJ716865 | KJ595435 | N/A |
| P. mercurial# | CBS 122443 | Macadamia integrifolia | KF853243 | DQ916363 | KF853236 | AB690666 | KJ595466 |
| P. middletonii# | CBS 528.74(VdPN) | Soil | N/A | AY598640 | AY598640 | AB362318 | KJ595457 |
| P. minus# | CBS 226.88 | Soil | HQ643696 | HQ643696 | HQ665168 | AB362320 | KJ595446 |
| P. monospermum# | CBS 158.73 (VdPN) | Soil | HQ643697 | HQ643697 | HQ643697 | KJ595350 | KJ595478 |
| P. montanum | CBS 111349 | Soil (Picea abies) | HQ643389 | HQ643389 | HQ643389 | KJ595410 | KJ595534 |
| P. multisporum | CBS 470.50 | Soil | AY598641 | AY598641 | AY598641 | AB362319 | KJ595455 |
| P. nagaii# | CBS 779.96 | Soil | AY598705 | AY598705 | AY598705 | KJ595402 | KJ595526 |
| P. nodosum# | CBS 102274 | Soil | N/A | HQ643709 | HQ665055 | KJ595407 | KJ595531 |
| P. nunn# | CBS 808.96 | Soil | AY598709 | AY598709 | AY598709 | AF196609 | DQ071325 |
| P. oedochilum# | CBS 292.37 | Unknown | AY598664 | AY598664 | AY598664 | AB108011 | EF408883 |
| P. okanoganense# | CBS 315.81 | Triticum aestivum | AY598649 | AY598649 | AY598649 | KJ595373 | KJ595498 |
| P. oligandrum# | CBS 382.34 (VdPN) | Viola sp. | AY598618 | AY598618 | AY598618 | KJ595381 | KJ595505 |
| P. oopapillum# | CBS 124053 | Cucumis sativus | N/A | FJ655174 | FJ655174 | KJ595431 | KJ595556 |
| P. ornacarpum | CBS 112350 | Soil | HQ643721 | HQ643721 | HQ643721 | KJ595411 | KJ595535 |
| P. ornamentatum | CBS 122665 | Soil | N/A | HQ643722 | HQ665117 | KJ595428 | KJ595552 |
| P. orthogonon# | CBS 376.72 | Zea mays | AY598710 | AY598710 | HQ665221 | KJ595379 | KJ595503 |
| P. oryzicollum# | Kr7 | soil | N/A | KX228072 | N/A | KX228125 | KX228108 |
| P. ostracodes# | CBS 768.73 (VdPN) | Soil | AY598663 | AY598663 | AY598663 | AB690668 | EF408880 |
| P. pachycaule# | CBS 227.88 | Soil | AY598687 | AY598687 | HQ665169 | KJ595362 | KJ595487 |
| P. paddicum# | CBS 698.83 | Triticum and Hordeum | AY598707 | AY598707 | AY598707 | JX397982 | JX397968 |
| P. paroecandrum# | CBS 157.64 (VdPN) | Soil | AY598644 | AY598644 | AY598644 | DQ071391 | DQ071332 |
| P. parvum | CBS 225.88 | Soil | AY598697 | AY598697 | AY598697 | AB362322 | KJ595445 |
| P. pectinolyticum | CBS 122643 | Soil | HQ643739 | HQ643739 | HQ643739 | N/A | KJ595469 |
| P. periilum# | CBS 169.68 (VdPN) | Soil | AY598683 | AY598683 | HQ665141 | N/A | KJ595444 |
| P. periplocum# | CBS 289.31 | Citrullus vulgaris | AY598670 | AY598670 | AY598670 | KJ595369 | KJ595494 |
| P. perplexum# | CBS 674.85 | Viciafaba | AY598658 | AY598658 | AY598658 | KJ595395 | KJ595519 |
| P. phragmitis# | CBS 117104 | Soil (Phragmites australis) | HQ643746 | HQ643746 | HQ665081 | AJ890351 | EU152854 |
| P. pleroticum | CBS 776.81 | Nymphyoides peltata | AY598642 | AY598642 | AY598642 | AB362321 | KJ595461 |
| P. plurisporium# | CBS 100530 | Agrostis | AY598684 | AY598684 | AY598684 | KJ595405 | KJ595529 |
| P. polare# | CBS 118203 | Sanionia uncinata | KJ716858 | AB299390 | KJ716859 | KJ595417 | KJ595542 |
| P. polymastum# | CBS 811.70 (VdPN) | Lactuca sativa | AY598660 | AY598660 | AY598660 | KJ595403 | KJ595527 |
| P. porphyrae# | CBS 369.79 (VdPN) | Porphyrayezoensis | AY598673 | AY598673 | AY598673 | KJ595377 | KJ595501 |
| P. phragmiticola | P56 | Soil | N/A | KC145165 | N/A | KC145166 | KC145167 |
| P. prolatum# | CBS 845.68 | Rhododendron sp. | AY598652 | AY598652 | AY598652 | AB362330 | KJ595462 |
| P. pyrioosporum | IRAN2382C | Soil | N/A | KT894052 | N/A | – | N/A |
| P. pyrilobum# | CBS 158.64 | Pinus radiata | AY598636 | AY598636 | AY598636 | KJ595349 | KJ595477 |
| P. radiosum | CBS 217.94 | Soil | N/A | AY598695 | HQ665156 | KJ595356 | N/A |
| P. recalcitrans# | CBS 122440 | Soil (Vitis vinifera) | N/A | DQ357833 | KJ716861 | KJ595423 | EF195143 |
| P. rishiriense | GUCC0007 | Aquatic | N/A | AB998878 | N/A | – | N/A |
| P. rhizo-oryzae# | CBS 119169 | Soil | HQ643757 | HQ643757 | HQ643757 | KJ595420 | KJ595545 |
| P. rhizosaccharum | CBS 112356 | Soil (Saccharum officinarum) | N/A | HQ643760 | HQ665072 | AB362323 | KJ595463 |
| P. rostratifingens# | CBS 115464 | Soil (Malus sp.) | HQ643761 | HQ643761 | HQ643761 | KJ595416 | KJ595541 |
| P. rostratum# | CBS 533.74 | Soil | AY598696 | AY598696 | AY598696 | KJ595388 | KJ595512 |
| P. salpingophorum# | CBS 471.50 (VdPN) | Lupinus angustifolius | AY598630 | AY598630 | AY598630 | KJ595384 | KJ595508 |
| P. schmitthenneri# | CBS 129726 | Glycine max | N/A | JF836869 | KJ716862 | JF895530 | KJ595470 |
| P. scleroteichum# | CBS 294.37 | Ipomoea batatas | AY598680 | AY598680 | AY598680 | KJ595370 | KJ595495 |
| P. segnitium | CBS 112354 | Soil | HQ643772 | HQ643772 | HQ643772 | KJ595412 | KJ595537 |
| P. selbyi# | CBS 129728 | Zea mays | N/A | JF836871 | KJ716863 | JF895532 | KJ595471 |
| P. senticosum# | CBS 122490 | Soil (forest) | HQ643773 | HQ643773 | HQ643773 | AB362317 | KJ595467 |
| P. solare | CBS 119359 | Phaseolus vulgaris | N/A | EF688275 | KJ716860 | KJ595421 | KJ595546 |
| P. spinosum# | CBS 275.67 (VdPN) | Compost | AY598701 | AY598701 | AY598701 | KJ595366 | KJ595491 |
| P. splendens# | CBS 462.48 (VdPN) | Unknown | AY598655 | AY598655 | AY598655 | AB512921 | AB512852 |
| P. stipitatum | DAOM 240293 | Soil | N/A | KJ716866 | KJ716866 | KJ595437 | KJ595560 |
| P. sukuiense | CBS 110030 | Soil | N/A | HQ643836 | HQ665059 | KJ595408 | KJ595532 |
| P. sulcatum# | CBS 603.73 | Daucus carota | AY598682 | AY598682 | HQ665281 | KJ595393 | KJ595517 |
| P. sylvaticum# | CBS 453.67 | Soil | AY598645 | AY598645 | AY598645 | KJ595383 | KJ595507 |
| P. takayamanum | CBS 122491 | Soil | HQ643854 | HQ643854 | HQ643854 | AB362315 | KJ595468 |
| P. tardicrescens# | LEV1534 | Turfgrass | N/A | HQ643855 | HQ643855 | KJ595439 | KJ595562 |
| P. torulosum# | CBS 316.33 (VdPN) | Grass | AY598624 | AY598624 | AY598624 | KJ595374 | KJ595499 |
| P. tracheiphilum# | CBS 323.65 | Lactuca sativa | N/A | AY598677 | HQ665207 | KJ595375 | N/A |
| P. ultimum var. sporangiiferum# | CBS 219.65 | Chenopodium album | AKYB02045405 | AY598656 | AY598656 | KJ595357 | KJ595482 |
| P. ultimum var. ultimum# | CBS 398.51 | Lepidium sativum | AY598657 | AY598657 | AY598657 | KJ595382 | KJ595506 |
| P. uncinulatum# | CBS 518.77 | Lactuca sativa | AY598712 | AY598712 | AY598712 | KJ595385 | KJ595509 |
| P. undulatum# | CBS 157.69 (VdPN) | Soil (Pinus sp.) | AY598708 | AY598708 | AY598708 | KJ595348 | KJ595476 |
| P. urmianum | IRAN2376C | Soil | N/A | KT894049 | N/A | – | N/A |
| P. utonaiense | UZ00769 | Aquatic | N/A | KJ995587 | KJ995600 | N/A | N/A |
| P. vanterpoolii# | CBS 295.37 | Triticum aestivum | AY598685 | AY598685 | AY598685 | KJ595371 | KJ595496 |
| P. vexans# | CBS 119.80(VdPN) | Soil | HQ643400 | HQ643400 | HQ643400 | GU133518 | EF426556 |
| P. viniferum# | CBS 119168 | Soil (Vitis sp.) | HQ643956 | HQ643956 | HQ643956 | KJ595419 | KJ595544 |
| P. violae# | CBS 159.64 (VdPN) | Soil | AY598706 | AY598706 | AY598706 | JX397980 | JX397966 |
| P. wohlseniorum | W15-2 | Aquatic | N/A | MH277978 | MH289800 | MH289798 | MH289799 |
| Pythiumsp. rooibos 2 | STE-U 7550 | Aspalathus linearis | N/A | JQ412777 | N/A | JQ412813 | JQ412789 |
Fig 2 Maximum likelihood of Pythium species based on the concatenated SSU, ITS, LSU, cox2 and tub2 regions. The maximumparsimonious dataset consisted of 528 constant, 71 parsimony-informative and 556parsimony-uninformativecharacters. The parsimony analysis of the data matrix resulted in the maximum of ten equally mostparsimonious trees with a length of 1637 steps (CI = 0.200, RI = 0.737, RC = 0.147, HI = 0.800). ML and MP bootstrap support values over 60% are indicated. Type strains are in bold and the 11 clades (A-K) are indicated. Scale bar indicates number of substitutions per site. The tree was rooted with Lagenidium giganteum (CBS 580.84) and Lagenidium sp. (DAOM 242348 and CBS 127283). Likelihood of the best scoring ML tree was -28453.969593. Estimated base frequencies were as follows: A = 0.264872, C = 0.163069, G = 0.213432, T = 0.358627; substitution rates AC = 1.219282, AG = 3.062456, AT = 3.113530, CG = 0.855790, CT = 4.379562, GT = 1.000000.


No Comments