27 Oct Nothophoma
Nothophoma Qian Chen & L. Cai, Stud. Mycol. 82: 212 (2015)
Background
Nothophoma was introduced by Chen et al. (2015b) by transferring five Phoma species. Species are saprobes and pathogens. In addition, to the phytopathogens, N. gossypiicola has been isolated from clinical samples of humans in the respiratory secretion of a patient with pneumonia and a human bronchial wash sample (Valenzuela-Lopez et al. 2018). Chethana et al. (2019) showed that the comparative pathogenicity of Nothophoma species is low when compared to other opportunistic pathogens. Some species grow on other fungi or occur in soil (Boerema et al. 2004, Aveskamp et al. 2009, 2010, Chen et al. 2015b;). Some Nothophoma species might be host-specific to a single plant genus or family (Aveskamp et al. 2010; Chen et al. 2015b). However, there is no study of host-specificity in Didymellaceae. Abdel-Wahab et al. (2017) identified 55 bioactive compounds from an endophyte, N. multilocularis. Of these, ten compounds showed strong antimicrobial activity in combination.
Classification – Ascomycota, Pezizomycotina, Dothideomycetes, Pleosporomycetidae, Pleosporales, Didymellaceae
Type species – Nothophoma infossa (Ellis & Everh.) Qian Chen & L. Cai
Distribution – Argentina, China, Italy, India, Korea, Netherlands, Spain, Tunisia, Ukraine, United States
Disease symptoms – brown spot of fruits, leaf spots, shoot canker, stem cankers
Leaf spot produced by Nothophoma anigozanthi is elliptical to circular and black. Nothophoma pruni and N. quercina develop small, dark red or purple pinpoint lesions (Chethana et al. 2019). Liu et al. (2018b) identified N. quercina infection on ornamental crab-apple. Symptoms on the trunk appear as warts, the periderm around warts can become cracked, and the bark is roughened with a scaly periderm. During dry weather, these cankers expand and coalesce (Liu et al. 2018b; Fig 1). Nothophoma quercina develops shoot necrosis, stem browning, and wilted leaves on Chaenomeles sinensis (Yun and Oh 2016). In Tunisia, shoot blights caused by N. quercina were observed with diffuse cankers with gummosis on buds (Triki et al. 2019).
Hosts— Has a wide range of hosts including Anigozanthos flavidus, Anigozanthus maugleisii, Arachis hypogaea, Chaenomeles sinensis, Gossypium sp., Fraxinu spennsylvanica, Malus micromalus, Microsphaera alphitoides, Olea europaea, Phellodendrona murense, Pistacia vera, Prunu savium, Prunus dulcis, Spiraea salicifolia, Quercus sp. and Ziziphus jujube (Babaahmadi et al. 2018; Chen et al 2015b, 2017; Chethana et al. 2019; Jianyu et al. 2016; Liu et al. 2018b; Moral et al. 2017, 2018; Soleimani et al. 2018; Triki et al. 2019; Valenzuela-Lopez et al. 2018; Yun and Oh 2016; Zhang et al. 2020).
Morphological based identification and diversity
This genus was introduced by Chen et al. (2015b) based on molecular data to delineate a more natural classification for the Ascochyta–Didymella–Phoma species complex (Chen et al. 2015b; Fig. 23). Species produce elongate, barrel-shaped, olivaceous brown chlamydospores in chains (Chen et al. 2015b). However, there is little morphological variation among species (Valenzuela-Lopez et al. 2018).
Fig. 1 Nothophoma quercina on Malus micromalus aMalus micromalus (Crab-Apple tree).b Canker on the trunk. c, d appearance of conidiomata on trunk. e longitudinal section through conidiomata. f cross-section of conidiomata g, h conidiogenous cells. i, j conidia. k upper view on PDA. l reverse view on PDA. Scale bars: d=1000 μm e–f = 50μm g–j 10 μm.
Molecular based identification and diversity
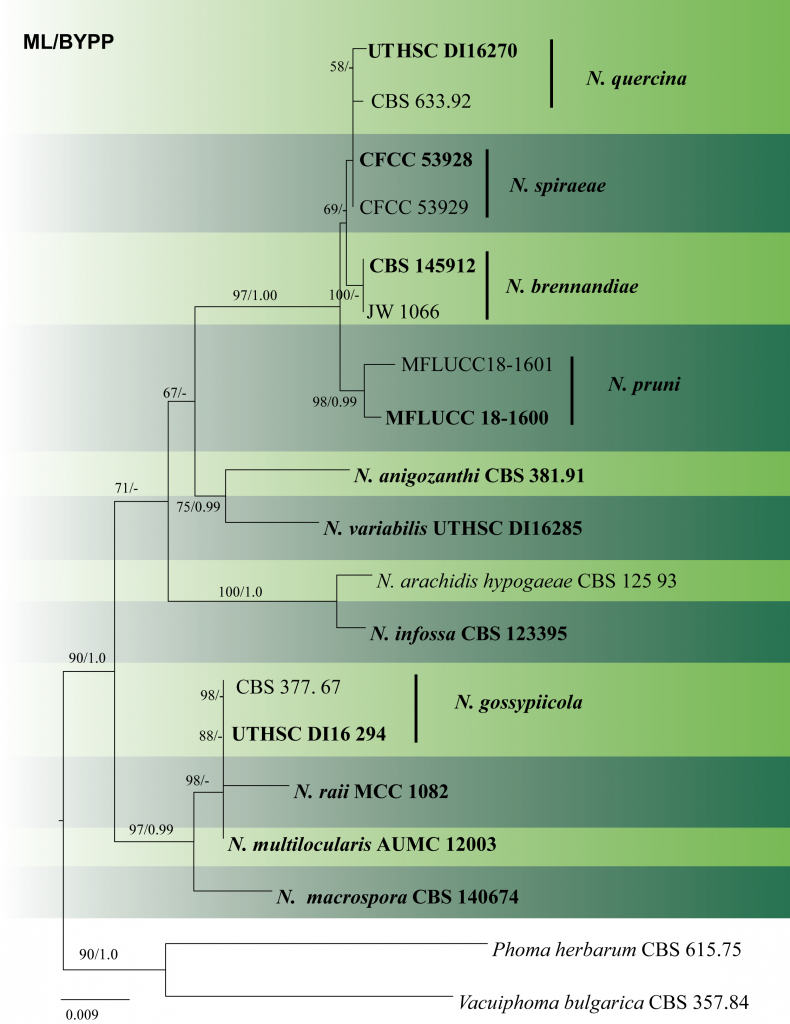
Species identification is based on multi-locus sequence phylogeny. Phylogenetic analyses of combined LSU, ITS, tub2 and rpb2 sequence data resulted in several new species being added to this genus by Chen et al. (2015b), Abdel-Wahab et al. (2017), Valenzuela-Lopez et al. (2018), Chethana et al. (2019), Marin-Felix et al. (2019) and Zhang et al. (2020). Here we provide an updated phylogenetic tree for this genus (Fig. 2).
Recommended genetic markers (genus level) – LSU, ITS
Recommended genetic markers (species level) – tub2, rpb2
Since the colony morphology and other morphological features in Didymellaceae often overlap, initial species identification is recommended with LSU and ITS sequence data using all type species in Didymellaceae. Once the genus is identified as Nothophoma, the phylogenetic analysis could be done with LSU, ITS, tub2, and rpb2 sequence data.
Accepted number of species – There are 12 species in Index Fungorum (2020) with DNA sequence data (Table 1).
References – Chen et al. 2015b, Abdel-Wahab et al. 2017, Valenzuela-Lopez et al. 2018, Chethana et al. 2019, Marin-Felix et al. 2019, Zhang et al. 2020 (morphology and phylogeny)
Table 1 DNA barcodes available for Nothophoma. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species | Isolate | LSU | ITS | tub2 | RPB2 |
| Nothophoma anigozanthi | CBS 381.91* | GU238039 | GU237852 | GU237580 | KT389655 |
| N. arachidis-hypogaeae | CBS 125.93 | GU238043 | GU237771 | GU237583 | KT389656 |
| N. brennandiae | CBS 145912* | MN823430 | MN823579 | MN824753 | MN824604 |
| JW 1066 | MN823429 | MN823578 | MN824752 | MN824603 | |
| N. gossypiicola | CBS 377.67 | GU238079 | GU237845 | GU237611 | KT389658 |
| UTHSC:DI16-294 | LN907437 | LT592943 | LT593012 | LT593082 | |
| N. infossa | CBS 123395 * | GU238089 | FJ427025 | FJ427135 | KT389659 |
| N. macrospora | CBS 140674 * | LN880537 | LN880536 | LN880539 | LT593073 |
| N. multilocularis | AUMC-12003* | KY996744 | – | – | – |
| N. pruni# | MFLUCC 18–1600 * | MH827028 | MH827007 | MH853671 | MH853664 |
| MFLUCC18–1601 | MH827026 | MH827005 | MH853669 | MH853662 | |
| N. quercina# | CBS 633.92 | EU754127 | GU237900 | GU237609 | KT389657 |
| UTHSC:DI16-270 | LN907413 | LT592929 | LT592998 | LT593067 | |
| N. raii | MCC 1082 * | – | MF664467 | MF664468 | – |
| N. spiraeae | CFCC 53928* | MN737828 | MN737833 | MN879295 | MN879292 |
| CFCC 53929 | MN737829 | MN737834 | MN879296 | MN879293 | |
| N. variabilis | UTHSC: DI16-285* | LN907428 | LT592939 | LT593008 | LT593078 |
Fig. 2 Phylogram generated from maximum likelihood analysis based on combined LSU, ITS, tub2 and rpb2 sequence data of Nothophoma species. Related sequences were obtained from GenBank. Seventeen strains are included in the combined sequence analyses. Phoma herbarum (CBS 615.75) and Vacuiphoma bulgarica (CBS 357.84) was used as the outgroup taxa. The best scoring RAxML tree with a final likelihood value of -5537.646741is presented. The matrix had 284 distinct alignment patterns, with 12.23% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.238395, C = 0.241637, G = 0.276596, T = 0.243371; substitution rates AC = 0.975188, AG = 4.004775, AT = 1.500008, CG = 0.519461, CT = 10.843965, GT = 1.000000; gamma distribution shape parameter a = 1.764918. ML bootstrap support value ≥50% and BYPP ≥0.95 are shown respectively near the nodes. Ex-type strains are in bold.


No Comments