27 Oct Thyrostroma
Thyrostroma Höhn., Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften Math.-naturw. Klasse Abt. I 120: 472 (1911)
Background
Thyrostroma belongs to Dothidotthiaceae of Pleosporales in Dothideomycetes, Ascomycota (Hongsanan et al. 2020). Thyrostroma was established by Höhnel (1911) and is typified by T. compactum. Thyrostroma had been treated as a synonym of Coryneum, Stegonsporium, Stigmina, and Thyrococcum, Thyrostromella and Wilsonomyces (Höhnel 1911; Morgan-Jones 1971; Sutton and Pascoe 1989; Sutton 1997; Index Fungorum 2020). Thyrostroma has been reported as the asexual morph of Dothidotthia based on the production of a hyphomycete state in culture (Ramaley 2005), however, there is no phylogenetic evidence to support this link. With new morphological information and phylogenetic analyses, Thyrostroma and Dothidotthia species were retained in separate genera (Crous et al. 2016; Marin-Felix et al. 2017; Senwanna et al. 2019). Thyrostroma species are pathogens, saprobes or endophytes associated with canker, dieback and leaf spots in terrestrial habitats (Yuan and Old 1990; Marin-Felix et al. 2017; Senwanna et al. 2019). Species of Thyrostroma have been recorded from various plants, however, host-specificity and pathogenic capacity of Thyrostroma has not yet been clarified.
Classification – Ascomycota, Pezizomycota, Dothideomycetes, Pleosporomycetidae, Pleosporales, Dothidotthiaceae
Type species – Thyrostroma compactum (Sacc.) Höhn
Distribution – Australia, Iran, Korea, Russia, USA, Uzbekistan
Disease Symptoms – Thyrostroma canker, dieback and leaf spots (Fig. 1a, b)
Hosts – Pathogens of Acanthophyllum sp., Astragalus sp., Capparis parvifiora, Celtis occidentalis, Cornus officinalis, Echinops sp., Elaeagnus angustifolia, Ephedra equisetina, Eucalyptus mannifera subsp. maculosa, Franseria sp., Halimodendron halodendron, Lycium barbarum, Morus alba, Robinia pseudoacacia, Sambucus caerulea, Styphnolobium japonicum, Tilia cordata, Ulmus pumila (Farr and Rossman 2020).
Morphological based identification and diversity
Thyrostroma species can be differentiated using conidial dimensions and septation in aged conidia and molecular phylogeny (Crous et al. 2016; Marin-Felix et al. 2017; Senwanna et al. 2019; Fig. 1). Senwanna et al. (2019) reported the sexual morph of Thyrostroma in T. ulmicola for the first time. The sexual morph is characterized by pseudothecial, immersed, erumpent or superficial, uniloculate or multiloculate ascostromata, globose to subglobose ascomata, a two-layered peridium, bitunicate, clavate asci, fusiform to ellipsoidal, 1-septate, ascospores.
Fig. 1 a, b. Symptoms on Ulmus pumila caused by Thyrostroma ulmicola (MFLU 16-1622); c, d Sporodochia on the host surface. e Section of sporodochium. f Conidiogenesis and conidiogenous cells. g–l Conidia. m Germinated conidium. Scale bars: d = 1000 µm, e = 200 µm, f–m = 30 µm.
Molecular based identification and diversity
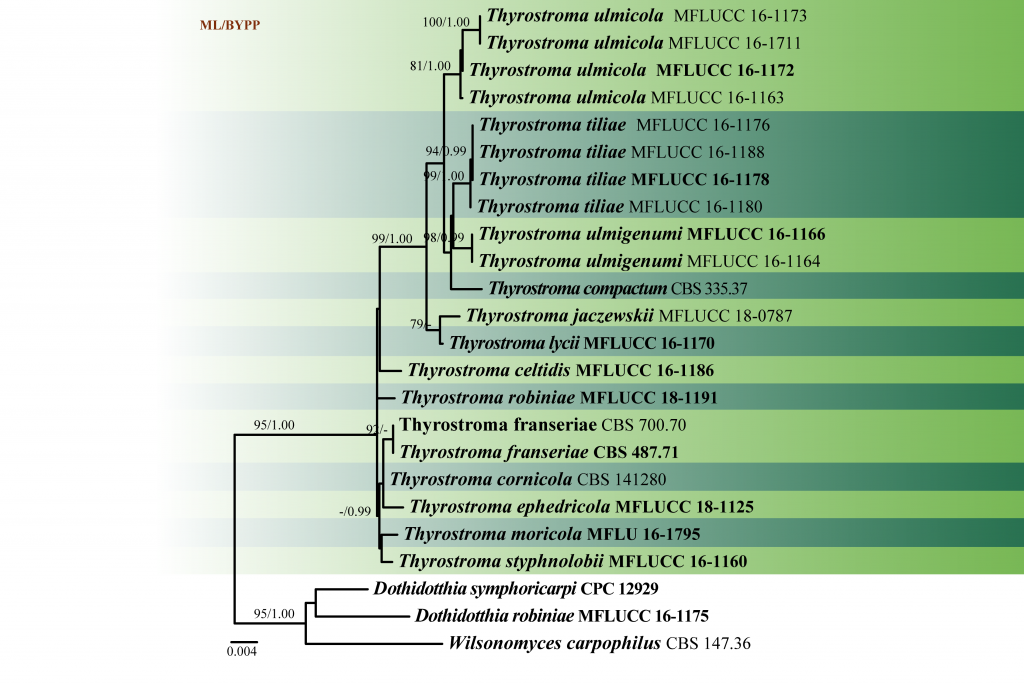
In the past, there has been no comprehensive phylogenetic study in Thyrostroma and consequently, its taxonomy was and still is mostly based on morphological characters. Based on LSU sequence data, Thyrostroma clustered in a well-supported clade within the Dothidotthiaceae (Marin-Felix et al. 2017; Crous et al. 2019). The asexual morph and sexual morph relationship were resolved by Senwanna et al. (2019) by molecular evidence. To achieve correct generic and species identification and taxonomic placement, phylogenetic studies using LSU, SSU, ITS, and tef1 were performed (Senwanna et al. 2019). This study reconstructs the phylogeny using a combined LSU, SSU, ITS, and tef1 sequence dataset (Fig. 2). The topology is in accordance with Marin-Felix et al. (2017), Senwanna et al. (2019) and Hyde et al. (2020b).
Recommended genetic marker (genus level) – LSU
Recommended genetic markers (species level) – ITS, tef1, rpb2 and tub2
LSU, ITS and tef1 are the common genetic markers used in the identification of Thyrostroma species. Combined LSU, SSU, ITS and tef1 genes provide a satisfactory resolution for resolving species. Based on the comparison of ITS and tef1gene regions, most species in Thyrostroma are not significantly different from one another, therefore, Senwanna et al. (2019) suggested that rpb2, tub2 are reliable genes for distinguishing species within Thyrostroma.
Accepted number of species – There are 27 epithets in Index Fungorum (2020), however only 13 species have DNA sequence data (Table 1).
References – Höhnel 1911, Yuan and Old 1990, Ramaley 2005 (morphology); Marin-Felix et al. 2017, Crous et al. 2016, 2019, Senwanna et al. 2019 (morphology and phylogeny).
Table 1 DNA barcodes available for Thyrostroma. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Isolate/Voucher no | LSU | SSU | ITS | tef1 |
| Thyrostroma compactum | CBS 335.37 | KY905664 | _ | KY905670 | KY905681 |
| T. cornicola | CBS 141280* | KX228300 | _ | KX228248 | KX228372 |
| T. ephedricola | MFLUCC 18-1125* | MK765854 | MK765853 | MK765855 | _ |
| T. franseriae | CBS 487.71* | KX228301 | _ | KX228249 | KY905680 |
| CBS 700.70 | KX228302 | _ | KX228250 | KY905682 | |
| T. jaczewskii | MFLUCC 18-0787 | MK765857 | MK765858 | MK765856 | _ |
| T. celtidis | MFLUCC 16-1186* | MK751822 | MK751767 | MK751732 | MK908022 |
| T. moricola | MFLU 16-1795* | MK751823 | MK751768 | MK751733 | MK908023 |
| T. lycii | MFLUCC 16-1170* | MK751824 | MK751769 | MK751734 | MK908024 |
| T. robiniae | MFLUCC 18-1191* | MK751825 | MK751770 | MK751735 | MK908025 |
| T. styphnolobii | MFLUCC 16-1160* | MK751826 | MK751771 | MK751736 | MK908026 |
| T. tiliae | MFLUCC 16-1176 | MK751827 | MK751772 | MK751737 | MK908027 |
| MFLUCC 16-1178* | MK751828 | MK751773 | MK751738 | MK908028 | |
| MFLUCC 16-1180 | MK751829 | MK751774 | MK751739 | MK908029 | |
| MFLUCC 16-1188 | MK751830 | MK751775 | MK751740 | MK908030 | |
| T. ulmicola | MFLUCC 16-1172* | MK751840 | MK751785 | MK751750 | MK908040 |
| MFLUCC 16-1173 | MK751841 | MK751786 | MK751751 | MK908041 | |
| MFLUCC 16-1163 | MK751834 | MK751779 | MK751744 | MK908034 | |
| MFLUCC 16-1711 | MK751839 | MK751784 | MK751749 | MK908039 | |
| T. ulmigenum | MFLUCC 16-1166* | MK751846 | MK751791 | MK751756 | MK908046 |
| MFLUCC 16-1164 | MK751845 | MK751790 | MK751755 | MK908045 |
Fig. 2 Phylogenetic tree generated by maximum likelihood analysis of LSU, SSU, ITS and tef1 sequence data of Thyrostroma species. Related sequences were obtained from GenBank. The tree was rooted with Dothidotthia robiniae (MFLUCC 16-1175), D. symphoricarpi (CPC 12929) and Wilsonomyces carpophilus CBS 147.36). Tree topology of the ML analysis was similar to the Bayesian analysis. The best scoring RAxML tree with a final likelihood value of -5556.187049 is presented. The matrix had 170 distinct alignment patterns, with 18.30% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243922, C = 0.240705, G = 0.270231, T = 0.245142; substitution rates AC = 3.570314, AG = 6.771467, AT 4.177691, CG = 1.603201, CT = 31.935571, GT = 1.000000; gamma distribution shape parameter α = 0.602378. Maximum likelihood bootstrap support values greater than 60% and Bayesian posterior probabilities ≥ 0.95 (BYPP) are indicated above the nodes. Ex-type (ex-epitype) and voucher strains are in bold.


No Comments