17 Sep Entyloma
Entyloma de Bary, Bot. Ztg. 32(7): 101(1874)
For synonyms see Index Fungorum (2018)
Background
Entyloma, known as the white smut fungus, was characterized by de Bary (1874). It forms teliospores with Tilletia-type basidia (holobasidia with apically produced basidiospores), and unique white coloration of the dense leaf spots caused by this pathogen. The genus was typified with E. microsporum from Ranunculus repens. The asexual genus Entylomella, described by Höhnel (1924), resembles Ramularia and was shown to be conspecific with Entyloma (Vánky 2012). Species of Entyloma have hyaline, globose, mostly smooth teliospores, embedded in the host tissue in the intercellular spaces of mesophyll cells. Prior to molecular phylogenetic studies, Entyloma species were described largely on the basis of morphology of spores and host associations. Some Entyloma species are important leaf pathogens of crops and ornamentals, including E. cosmi on Cosmos bipinnatus (Lutz and Piątek 2016), E. dahliae on Dahlia sp. (Fox 2014), E. eryngii-alpini on Eryngium alpinum (Vánky 2009; Savchenko et al. 2014), E. fuscum on Papaver sp. (Vánky 2012), E. gaillardianum on Gaillardia sp. (Glawe et al. 2010; Savchenko et al. 2012; Vánky 2012) and E. helianthi on Helianthus annuus (Rooney-Latham et al. 2017).
Classification – Exobasidiomycetes, Exobasidiomycetidae, Entylomatales, Entylomataceae
Type species – Entyloma microsporum J. Schröt., Fungi europ. Exsicc.: no.1872 (1874)
Distribution – Worldwide
Disease Symptoms – Leaf spot
Amphigenous circular pale green spots appear at the beginning with light to dark brown spots in the center (Garibaldi et al. 2018).
Hosts – Species are e host specific and found on a variety of dicotyledonous hosts with more than 80% of the species occurring on asterids and ranunculoids. Entyloma species have also been recorded from other plant families, with major ones being Apiaceae, Fabaceae, Papaveraceae, Primulaceae, Saxifragaceae, Scrophulariaceae, and Solanaceae (Vánky 2012).
Morphological based identification and diversity
Entyloma was formerly a broad genus that included almost all of the species of smut fungi with solitary teliospores produced intercellularly in host tissue. With the advent of DNA sequence data, former Entyloma species occurring on monocots were transferred to several genera in the order Georgefischeriales (Begerow et al. 1997, 2002; Bauer et al. 2001, 2005).The number of species recognized within the genus has varied depending upon the taxonomic concept used. Morphology-based classification of Entyloma species was proposed by Savile (1947). The species were based exclusively on spore size and asexual morph. The adoption of a morphological concept dramatically decreased the number of recognized species and synonymized those with identical morphology found on the same host family. The application of this concept is challenging as it is mostly based on very simple characters (sorus morphology, size and colour of the spores, and thickness and surface of spore walls) that often overlap. Vánky in his European and World monographs of smut fungi (1994, 2012) proposed a narrower concept based on both morphological data and host-specificity. Subsequent molecular studies on the genus supported the idea that the species of Entyloma are restricted to the hosts within a single genus or species, hence the taxonomic concept proposed by Vánky proved to be more evolutionary correct than that of Savile (Begerow et al. 2002; Savchenko et al. 2014, 2015; Lutz and Piątek 2016; Rooney-Latham et al. 2017; Kruse et al. 2018). However, many of the host specific species are known only from limited collections, and some have been collected only once. Therefore, the information on host specificity may change with further collections.
Identification of Entyloma using morphological species criteria is not always accurate, and should include molecular and host data. However, lack of DNA sequences for more than half of the known species is a problematic issue for molecular identification of the Entyloma species.
Sori and spore morphology are the primary characters to identify species within this genus (Savchenko et al. 2014, 2015; Vánky 1994, 2012). However, a lot of species of Entyloma are morphologically indistinguishable and the taxonomic position of the host should be considered as another character.
Molecular based identification and diversity
The first phylogenetic analysis for Entyloma by Begerow et al. (2002) used ITS and LSU regions as separate genetic markers and showed that the highest resolution resulted from the analysis of ITS sequence data (Begerow et al. 2002). That study grouped all Entyloma species into two major clades, one with the species occurring on ranunculids, and another with the species from asterids. Later studies on the molecular systematics of Entyloma, focused on revealing phylogenetic relationships among particular groups of species and species complexes, supported this grouping (Savchenko et al. 2014, 2015; Lutz and Piątek 2016; Rooney-Latham et al. 2017; Kruse et al. 2018). Studies applying these tools are revealing significantly greater diversity on some hosts than was previously realized.
Most taxonomic studies on Entyloma using molecular data have employed ITS rDNA phylogenies, and this single marker has been shown to be reliable in species delimitation if used in combination with morphological and host data (Savchenko et al. 2014, 2015; Lutz and Piątek 2016; Rooney-Latham et al. 2017). A phylogenetic tree of the genus Entyloma based on ITS data is presented. Based on this phylogenetic tree species of Entyloma on asterids are in two major clades. Both clades include species parasitizing Asteraceae, and plants from other host families, including several members of Ranunculids and Euasterids. The results of this work are similar to previous studies on phylogenetics of Entyloma (Begerow et al. 2002; Savchenko et al. 2014, 2015; Lutz and Piątek 2016; Rooney-Latham et al. 2017).
Recommended genetic markers (Genus level) – ITS
Recommended genetic markers (Species level) – ITS
Identification of Entyloma species is complicated because of its simple morphology and often small genetic differences between species. The combination of morphology, genetic information, and host data is sufficient for identification of particular species.
Accepted number of species: Currently, there are 458 Entyloma names listed in Index Fungorum (2018), but only about 170 were accepted in the world monograph of smut fungi by Vanky (2012) and subsequent publications dealing with this genus (Denchev et al. 2013; Savchenko et al. 2014, 2015, 2017; Rooney-Latham et al. 2017).
References: Begerow et al. 2002, Vánky 2012, Savchenko et al. 2014, 2015, Lutz and Piątek 2016, Rooney-Latham et al. 2017, Kruse et al. 2018 (morphology and phylogeny), Vánky 2012 (morphology, host specificity)
Table Entyloma Details of the isolates used in the phylogenetic analyses. Type species for the genus is in bold and marked with an asterisk.
| Species | Host | GenBank Accession Number (ITS) |
| Entyloma arnicale | Arnica montana | AY854964 |
| E. arnicale | Arnica montana | AY854965 |
| E. arnoseridis | Arnoseris minima | AY081017 |
| E. australe | Physalis cordata | AY081019 |
| E. bidentis | Bidens pilosa | AY081020 |
| E. browalliae | Browallia americana | AY081021 |
| E. browalliae | Browallia americana | AY854962 |
| E. calceolariae | Calceolaria chelidonioides | AY081022 |
| E. calendulae | Calendula officinalis | AY081023 |
| E. carmeli | Eryngium falcatum | KF310892 |
| E. carmeli | Eryngium falcatum | KF310893 |
| E. chrysosplenii | Chrysosplenium alternifolium | AY081024 |
| E. chrysosplenii | Chrysosplenium alternifolium | AY854960 |
| E. comaclinii | Comaclinium montanum | AY081025 |
| E. compositarum | Parthenium hysterophorus | AY081026 |
| E. cosmi | Cosmos bipinnatus | KJ728761 |
| E. corydalis | Corydalis bulbosa | AY081027 |
| E. costaricense | Viguiera sp. | AY081028 |
| E. dahliae | Dahlia variabilis | AY854975 |
| E. delileae | Delilea biflora | AY081030 |
| E. diastateae | Diastatea micrantha | AY081031 |
| E. diastateae | Diastatea micrantha | AY854974 |
| E. doebbeleri | Dahlia imperialis | AY081032 |
| E. doebbeleri | Dahlia imperialis | AY854973 |
| E. eryngii | Eryngium campestre | AY081033 |
| E. eryngii | Eryngium campestre | KF310897 |
| E. eryngii | Eryngium campestre | KF310896 |
| E. eryngii-cretici | Eryngium creticum | KF310894 |
| E. eryngii-cretici | Eryngium creticum | KF310895 |
| E. eryngii-plani | Eryngium planum | AY081034 |
| E. eschscholziae | Eschscholzia california | KC456226 |
| E. fergussonii | Myosotis sp. | AY854970 |
| E. fergussonii | Myosotis palustris | AY854971 |
| E. ficariae | Ranunculs ficaria | JQ586199 |
| E. gaillardianum | Gaillardia aristata | AY081037 |
| E. gaillardianum | Gaillardia aristata | AY854968 |
| E. gaillardianum | Gaillardia aristata | GU117108 |
| E. guaraniticum | Bidens pilosa | AY081038 |
| E. hieracii | Hieracium sylvaticum | AY081039 |
| E. hieracii | Hieracium lachenalii | AY854967 |
| E. hieracii | Hieracium murorum | EU233810 |
| E. lobeliae | Lobelia laxiflora | AY081042 |
| E. madiae | Madia gracilis | AY081043 |
| E. magocsyanum | Tordylium cordatum | KF310891 |
| E. matricariae | Tripleurospermum perforatum | AY081044 |
| E. matricariae | Tripleurospermum perforatum | AY854979 |
| E. microsporum* | Ranunculus repens | AY081045 |
| E. polysporum | Ambrosia artemisifolia | AY081046 |
| E. scandicis | Scandix verna | KF447774 |
| E. scandicis | Scandix verna | KF447775 |
| E. zinniae | Zinnia peruviana | AY081049 |
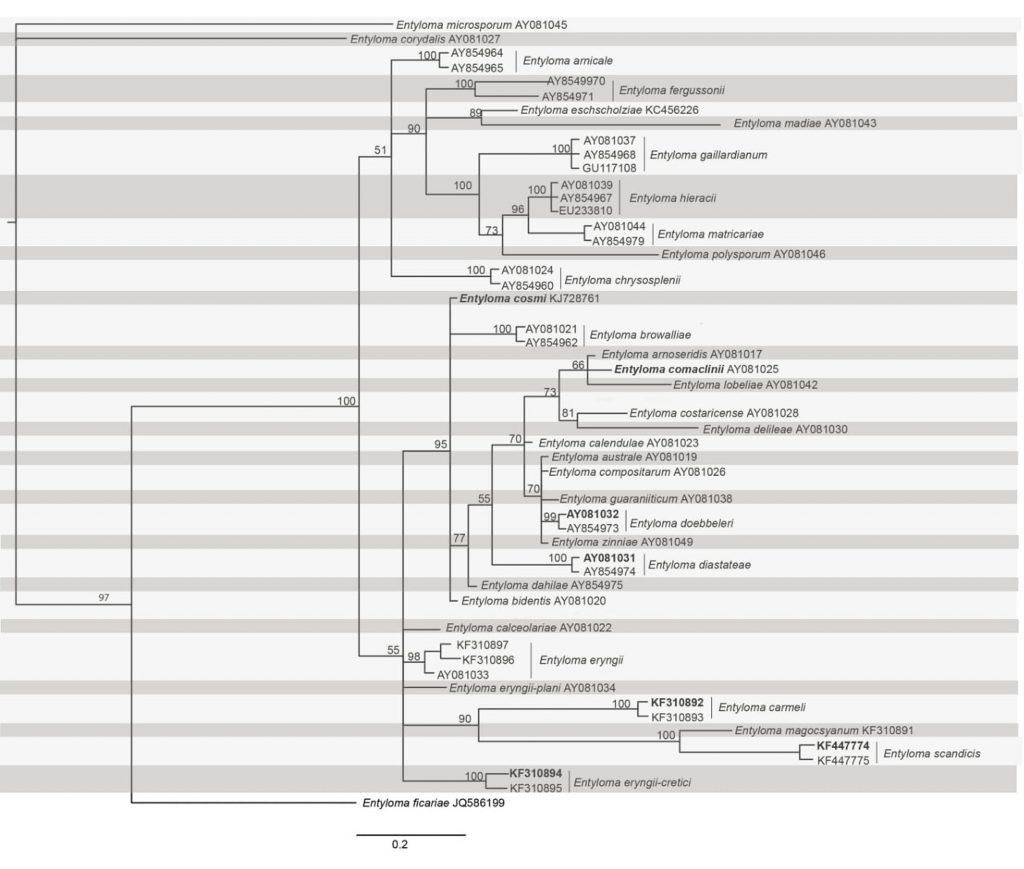
Fig Phylogenetic tree generated from Bayesian inference based on ITS nucleotide sequence data of Entyloma species. Related sequences were obtained from GenBank. The ITS alignment included 51 sequences comprising 629 characters (including gaps). Parsimony analysis revealed that 497 characters are constant and of the variable characters 87 are parsimony-uninformative and 104 are parsimony informative. The parsimony analysis yielded 35 equally parsimonious trees, and the strict consensus tree of all equally parsimonious trees was used. The different runs of Bayesian phylogenetic analyses yielded consistent topologies. Bayesian posterior probabilities greater than 0.5 are indicated near the nodes. Numbers on branches are estimates for PPs >0.5. The tree is rooted with Entyloma microsporum. The type specimens are in bold.

No Comments