24 Oct Dothidotthia
Dothidotthia Höhn., Berichte der Deutschen Botanischen Gesellschaft 36: 312 (1918)
Background
Dothidotthia was assigned to Botryosphaeriaceae, because of its coelomycetous asexual morph, and characteristic peridium, pseudoparaphyses and asci (Barr 1989). Ramaley (2005) reported that Thyrostroma is the asexual morph of Dothidotthia based on the production of hyphomycetes in culture. Phillips et al. (2008), introduced a new family Dothidotthiaceae to accommodate Dothidotthia and considered Thyrostroma as the asexual morph of Dothidotthia. However, the links between the sexual and asexual morphs are not supported by molecular evidence. Recent molecular and morphology studies (Marin-Felix et al. 2017; Crous et al. 2019; Senwanna et al. 2019), based on a taxon sampling of current species indicates that Dothidotthia does not cluster near Thyrostroma. Thus, Dothidotthia is a distinct genus.
Classification – Ascomycota, Pezizomycotina, Dothideomycetes, Pleosporomycetidae, Pleosporales, Dothidotthiaceae
Type species – Dothidotthia symphoricarpi (Rehm) Höhn.
Distribution – in both temperate and tropical countries (Italy, Russia, Thailand, Ukraine and the USA)
Disease symptoms – species cause canker, dieback and leaf spot diseases on twig, branch, bark and leaf
Hosts – Pathogens of Acer negundo, Diapensia lapponica, Fendlera rupicola, Euonymus alatus, Robinia pseudoacacia, Verbena asparagoides (Barr 1989; Farr and Rossman 2019; Index Fungorum 2020).
Morphological based identification and diversity
In previous studies, the asexual morphs of Dothidotthia have been reported as Thyrostroma (Ramaley 2005), however, phylogenetic analyses indicated that Dothidotthia can be separated from Thyrostroma (Marin-Felix et al. 2017; Crous et al. 2016; Senwanna et al. 2019). Dothidotthia is characterized by fusiform to obclavate or obpyriform, 0–3-transversely septate conidia and a sexual morph with clavate, short pedicellate asci, ellipsoid, 1-septate ascospores (Fig. 1). The sexual morphs of Dothidotthia and Thyrostroma have similar morphological characteristics in shape and overlapping dimensions of asci and ascospores (Barr 1989; Ramaley 2005; Phillips et al. 2008; Hyde et al. 2013; Senwanna et al. 2019). However, Dothidotthia can be differentiated from Thyrostroma by peridium structure and conidial morphology and molecular phylogeny (Senwanna et al. 2019). Crous et al. (2019) introduced Neodothidotthia to accommodate N. negundinicola and Dothidotthia aspera was synonymized under N. negundinis based on analysis of LSU sequence data. However, Senwanna et al. (2019) showed that Neodothidotthia negundinicola and N. negundinis group with D. robiniae and D. symphoricarpi (type species). Furthermore, the conidial morphology of Neodothidotthia is similar to Dothidotthia symphoricarpi (Pseudotthia symphoricarpi) and D. robiniae (Phillips et al. 2008; Zhang et al. 2012; Crous et al. 2019; Senwanna et al. 2019). Therefore, Neodothidotthia had been treated as a synonym of Dothidotthia.
Fig. 1 Dothidotthia robiniae (MFLU 16-1704). a, b Sporodochia on the host surface. c Vertical section of sporodochium. d Conidiogenesis. e, g Conidia attached with the conidiogenous cells. f, h Conidia. i Germinated conidium. Scale bars: b = 1000 µm, c = 200 µm, d–i = 30 µm.
Molecular based identification and diversity
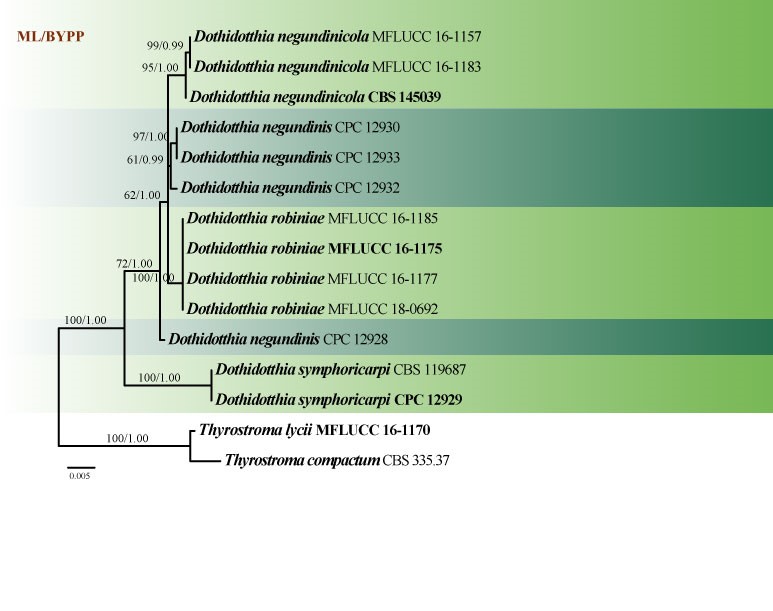
Dothidotthia species can be separated from Thyrostroma based on LSU sequence data (Marin-Felix et al. 2017; Crous et al. 2019). Multigene phylogenetic analyses of a combined LSU, SSU, ITS and tef1 dataset for Dothidotthia is presented in this study, which is similar to Senwanna et al. (2019) (Fig. 2).
Recommended genetic markers (genus level) – LSU, SSU
Recommended genetic markers (species level) – ITS, tef1, rpb2 and tub2
Accepted number of species – There are 14 epithets listed in Index Fungorum (2020), however only four species have DNA molecular data (Table 1).
References – Barr 1989, Ramaley 2005 (morphology); Phillips et al. 2008, Zhang et al. 2012, Hyde et al. 2013, Marin-Felix et al. 2017, Crous et al. 2019, Senwanna et al. 2019 (morphology and phylogeny)
Table 1 DNA barcodes available for Dothidotthia. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Isolate no | LSU | SSU | ITS | tef1 |
| Dothidotthia negundinicola | CBS 145039* | MK442537 | _ | MK442597 | _ |
| MFLUCC 16-1157 | MK751815 | MK751760 | MK751725 | MK908015 | |
| MFLUCC 16-1183 | MK751816 | MK751761 | MK751726 | MK908016 | |
| D. negundinis | CPC 12930 | EU673274 | EU673226 | MK442599 | _ |
| CPC 12932 | EU673275 | EU673227 | MK442600 | _ | |
| CPC 12933 | EU673276 | EU673228 | MK442601 | _ | |
| D. robiniae | MFLUCC 16-1175* | MK751817 | MK751762 | MK751727 | MK908017 |
| MFLUCC 16-1177 | MK751818 | MK751763 | MK751728 | MK908018 | |
| MFLUCC 16-1185 | MK751819 | MK751764 | MK751729 | MK908019 | |
| MFLUCC 18-0692 | MK751821 | MK751766 | MK751731 | MK908021 | |
| D. symphoricarpi | CPC 12929* | EU673273 | EU673224 | _ | _ |
| CBS 119687 | MH874618 | _ | MH863064 | _ |
Fig. 2 Phylogenetic tree generated by ML analysis of LSU, SSU, ITS and tef1 sequence data of Dothidotthia species. Related sequences were obtained from GenBank. The tree was rooted with Thyrostroma compactum (CBS 335.37) and T. lycii (MFLUCC 16-1170). Tree topology of the ML analysis was similar to the Bayesian analysis. The best scoring RAxML tree with a final likelihood value of -5116.933762 is presented. The matrix had 115 distinct alignment patterns, with 25.41% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.245094, C = 0.237101, G = 0.269739, T = 0.248067; substitution rates AC = 3.925871, AG = 7.445430, AT = 2.745308, CG = 2.728664, CT = 20.049514, GT = 1.000000; gamma distribution shape parameter α = 0.790240. Maximum likelihood bootstrap support values greater than 60% and BYPP probabilities ≥0.95 are indicated above the nodes. Ex-type (ex-epitype) and voucher strains are in bold.


No Comments