17 Sep Fulvifomes
Fulvifomes Murrill, North. Polyp.: 49(1914)
For synonyms see Index Fungorum (2018)
Background
Fulvifomes was described by Murrill (1914) and it is typified by F. robiniae (Murrill) Murrill. Originally, Fulvifomes was characterized by “hymenophore large, perennial, epixylous, sessile, ungulate or applanate; surface sulcate, usually anoderm and often rough or rimose; context woody or punky, brown, rarely dark-red; tubes brown, cylindric, stratose, usually thick- walled; spores smooth, ferruginous or fulvous” and its species were reported as growing on living hosts (Fagus, Juniperus, Quercus, Ribes, Robinia) (Murrill 1914). Wagner and Fischer (2001, 2002) redefined the genus as saprobic on deciduous wood, with resupinate, effused-reflexed or pileate, perennial basidiomata, dimitic hyphal system, ellipsoid, yellowish, IKI- basidiospores and lack of sterile elements like setae. Later, new species were described (Hattori et al. 2014; Zhou 2014, 2015; Ji et al. 2017; Salvador-Montoya et al. 2018) and a new definition of Fulvifomes was provided to include species with substipitate basidiomata with contracted base, solitary or imbricate, corky to woody hard, with pileal surface tomentose or glabrous, with or without a crust; context homogenous or duplex; hyphal system monomitic or dimitic; basidiospores subglobose to ellipsoid, yellowish to brown, fairly thick- to thick-walled, CB- or CB+; and occurring also on gymnosperms. Some species seem to be restricted to specific hosts, while others appear to be generalists (Murrill 1914; Larsen et al. 1985; Larsen and Cobb-Poule 1990; Sakayaroj et al. 2012; Hattori et al. 2014; Zhou 2015; Ji et al. 2017), but most host information is based on only a few collections.
Classification – Agaricomycetes, incertae sedis, Hymenochaetales, Hymenochataceae
Type species – Fulvifomes robiniae (Murril) Murrill, North. Polyp.: 49(1914)
Distribution – Cosmopolitan
Disease Symptoms – White pocket rot
Species of Fulvifomes develop a white pocket rot in their hosts (Larsen et al. 1985; Holmquist 1990). The pockets are irregular and appear to be interconnected by radially oriented decayed areas. The masses of fungal hyphae are white and the wood between decay pockets has a friable and crumbly texture (Larsen et al. 1985).
Hosts – Acacia sp., Amburana cearensis, Anadenanthera colubrina, Apuleia leiocarpa, Aspidosperma quebracho-blanco, Bombax sp., Casuarina sp., Cedrela sp., Fagus sp., Gliricidia sp., Juniperus sp., Krugiodendron sp., Mimozyganthus carinatus, Mora gonggrijpii, Prosopis sp., Parapiptadenia rigida, Patagonula americana, Peltophorum dubium, Prunus subcoriacea, Quercus sp., Ribes sp., Robinia sp., Schinus sp., Shorea sp., Xylosma venosa, Xylocarpus sp.and Ziziphus mistol (Wright and Blumenfeld 1984; Urcelay et al. 1999; Robledo and Urcelay 2009).
Morphological based identification and diversity
Species of Fulvifomes were previously mostly identified as species of Phellinus sensu lato. (Gilbertson and Ryvarden 1987; Larsen and Cobb-Poule 1990; Ryvarden and Gilbertson 1994; Ryvarden 2004; Dai 2010). However, when Wagner and Fischer (2001, 2002) studied the poroid Hymenochaetaceae, they resurrected several genera placed under synonymy with Phellinus, among them, Fulvifomes.
Fulvifomes can be identified by the macro- and micromorphology of its basidiomata. Identification of species is not always accurate when only using morphological characters and the use of molecular data has been shown to be very useful.
Molecular based identification and diversity
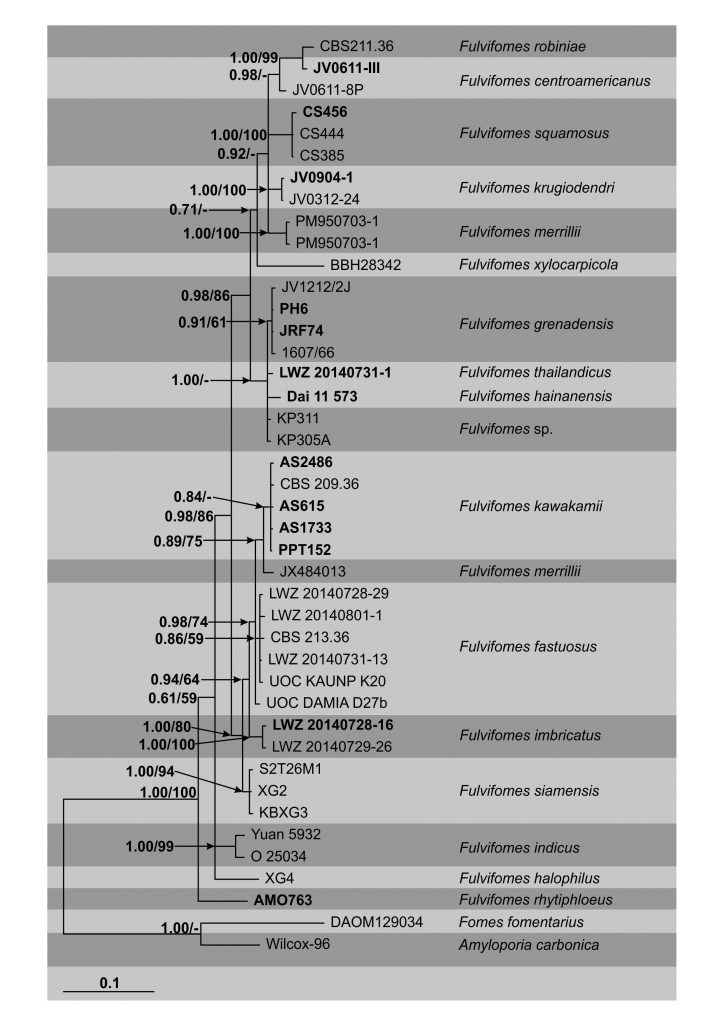
The first phylogenetic analysis for Fulvifomes was carried out by Wagner and Fischer (2001, 2002) when studying the poroid Hymenochaetaceae. The authors used sequence data from the LSU rDNA and recovered Fulvifomes among Phellinus species. Latter, Zhou (2014, 2015), Ji et al. (2017) and Salvador-Montoya et al. (2018) described new species using both LSU rDNA and ITS sequence data, in separate or combined analyses. Here we reconstruct the phylogeny of Fulvifomes based on the combined analyses of ITS and LSU rDNA sequence data. This tree includes a reference sequence of the type species of the genus, collected in the same country and on the same host, and the sequence of the type of newly described F. squamosus Salvador-Montoya & Drechsler-Santos (Salvador-Montoya et al. 2018) and provides the first sequence of F. rhytiphloeus (Mont.) Camp.-Sant. & Robledo. Additionally, the tree implies the absence in the Americas of F. fastuosus (Lév.) Bondartseva & S. Herrera, F. merrillii (Murrill) Baltazar & Gibertoni and F. nilgheriensis (Mont.) Bondartseva & S. Herrera, whose type localities are in Asia, and also implies the wider distribution of F. kawakamii (M.J. Larsen, Lombard & Hodges) T. Wagner & M. Fisch., previously thought to be endemic to Hawai’i (EUA, Larsen et al. 1985) (99% similarity and 89% query cover with Fu. kawakamii AY059028 from the type locality, LSU only), and probably identified as Fu. nilgheriensis (CBS 209.36) or Fu. fastuosus (other specimens collected in the Americas).
Recommended genetic markers (Genus level) – LSU
Recommended genetic markers (Species level) – ITS, TEF1-α and RPB2 as additional markers
Matheny et al. (2007) studied the level of resolution of TEF1-α and RPB2 in phylogeny of Basidiomycota, concluding that RPB2 is more efficient to resolve both higher and lower clades, while TEF1-α is better to solve the phylogeny in high taxonomic levels. Considering rDNA markers, LSU rDNA is used for genera delimitation, while ITS rDNA is used to delimit species (James et al. 2006; Matheny et al. 2007; Öpik et al. 2010; Schoch et al. 2012). In Hymenochataceae, these two regions are extensively used in many phylogenies, to discriminate taxa in the family (Wagner and Fischer, 2001, 2002; Larsson et al. 2006). In Fulvifomes, many studies used sequences from ITS and LSU rDNA as markers (Zhou 2014; 2015; Ji et al. 2017; Salvador-Montoya et al. 2018). However, TEF1-α and RPB2 are also being used for delimitation of taxa of poroid Hymenochaetaceae, for instance in Fomitiporia and Phellinus (Amalfi et al. 2010, 2014; Campos-Santana et al. 2014, 2016; Chen and Cui 2017; Morera et al. 2017).
Accepted number of species: There are 58 species epithets in Index Fungorum (2018) under this genus. However, only 24 are accepted.
References: Wagner and Fischer 2001, 2002 (phylogeny), Hattori et al. 2014 (morphology), Zhou 2014, 2015, Ji et al. 2017, Salvador-Montoya et al. 2018 (morphology, phylogeny).
Table Fulvifomes. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an *
| Species | Voucher | LSU | ITS |
| F. centroamericanus | JV0611-8P | – | KX960757 |
| F. centroamericanus | JV0611-III* | KX960764 | KX960763 |
| F. fastuosus | LWZ 20140731-13 | KR905668 | KR905674 |
| F. fastuosus | CBS 213.36 | AY059057 | AY558615 |
| F. fastuosus | LWZ 20140801-1 | KR905669 | KR905675 |
| F. fastuosus | UOC DAMIA D27b | – | KJ206286 |
| F. fastuosus | LWZ 20140728-29 | – | KR905673 |
| F. fastuosus | UOC KAUNP K20 | – | KR867659 |
| F. grenadensis | JV1212/2J | – | KX960756 |
| F. grenadensis | 1607/66 | KX960758 | |
| F. grenadensis | JRF74 | MH048087 | MH048097 |
| F. grenadensis | PH6 | MH048086 | MH048096 |
| F. hainanensis | Dai 11 573* | JX866779 | KC879263 |
| F. halophilus | XG4 | JX104752 | JX104705 |
| F. imbricatus | LWZ 20140728-16* | KR905670 | KR905677 |
| F. imbricatus | LWZ 20140729-26 | KR905671 | KR905679 |
| F. indicus | O 25034 | KC879259 | KC879262 |
| F. indicus | Yuan 5932 | JX866777 | KC879261 |
| F. kawakamii | PPT152 | MH048085 | MH048095 |
| F. kawakamii | AS1733 | MH048083 | MH048093 |
| F. kawakamii | AS615 | MH048082 | MH048092 |
| F. kawakamii | AS2486 | MH048084 | MH048094 |
| F. kawakamii | CBS 428.86 | AY059028 | – |
| F. krugiodendri | JV0904-1* | KX960765 | KX960762 |
| F. krugiodendri | JV0312-24 | KX960760 | KX960766 |
| F. merrillii | PM950703-1 | – | EU035313 |
| F. merrillii | PM950703-1 | – | EU035310 |
| F. merrillii | – | – | JX484013 |
| F. nilgheriensis | CBS 209.36 | AY059023 | AY558633 |
| F. rhytiphloeus | AMO763 | MH048081 | MH048091 |
| F. robiniae | CBS 211.36 | – | AY558646 |
| F. siamensis | XG2 | JX104756 | JX104709 |
| F. squamosus | CS456* | MF479266 | MF479267 |
| F. squamosus | CS385 | MF479265 | MF479268 |
| F. squamosus | CS444 | MF479264 | MF479269 |
| F. thailandicus | LWZ 20140731-1* | KR905665 | KR905672 |
| F. xylocarpicola | BBH 28342 | JX104723 | JX104676 |
| Fulvifomes sp. | S2T26M1 | JX104754 | JX104707 |
| Fulvifomes sp. | KBXG3 | – | JX104706 |
| Fulvifomes sp. | KP311 | – | KP658651 |
| Fulvifomes sp. | KP305A | – | KP658646 |
Fig. Phylogenetic tree generated by Bayesian inference (BI) of combined ITS and LSU rDNA sequence data of Fulvifomes species. Forty samples are included in the analyses, which comprise 1224 characters including gaps. Tree was rooted with Fomes fomentarius (DAOM129034) and Amyloporia carbonica (Wilcox-96). Tree topology of the BI was similar to the maximum likelihood (ML) analysis (Figure not shown). The matrix had 1021 phylogenetic informative sites (83, 42%). Estimated base frequencies were as follows; A = 0.241, C = 0.208, G = 0.280, T = 0.271; substitution rates AC = 0.768, AG = 4.476, AT = 1.569, CG = 1.133, CT = 8.068, GT = 1.000; gamma distribution shape parameter α = 0.246. Bayesian posterior probabilities and ML bootstrap values ≥50% are shown respectively near the nodes. The scale bar indicates 0.05 changes. Sequences generated in this study and of the types are in bold.

No Comments