26 Oct Fomitopsis
Fomitopsis P. Karst., Meddn Soc. Fauna Flora fenn. 6: 9 (1881)
Background
Fomitopsis was established by Karsten (1881) based on four species, with F. pinicola as the generic type (Murrill 1903; Donk 1960). The genus has a cosmopolitan distribution and comprises species causing brown rot on both living and dead trees (Han et al. 2016). Fomitopsis species also contribute to the decomposition of coarse woody debris in forest communities (Gilbertson 1980; Haight et al. 2019). There are certain instances of their pathogenic role in orchards of cultivated species where they cause heart rot on Citrus (Roccotelli et al. 2014) and Prunus species (Adaskaveg 1993). A Fomitopsis sp. was also recorded in oil palm (Elaeis guineensis) as an endophyte (Rungjindamai et al. 2008; Pinruan et al. 2010).
Classification – Basidiomycota, Agaricomycetes, Incertae sedis, Polyporales, Fomitopsidaceae
Type species – Fomitopsis pinicola (Sw.) P. Karst.
Distribution – Worldwide
Disease symptoms – Fomitopsis causes brown cubical rot on both living and dead trees (Mounce 1929). The basidiospores can be dispersed by wind, or by vectors such as bark beetles (Castello et al. 1976; Pettey and Shaw 1986; Lim et al. 2005; Persson et al. 2011; Jacobsen et al. 2017; Vogel et al. 2017). Upon infecting standing trees, stumps, or logs through wounds, or through the tunnels of penetrating vectors, the fungus establishes itself in the xylem (Mounce 1929). The growth rate of Fomitopsis species in the substrata can differ depending on their ecological requirements (Markovic et al. 2011; Haight et al. 2019). When the decay starts, the wood turns yellowish-brown, which later splits into cubical fragments. The colour is generally lighter in case of F. pinicola than other agents of brown rot decay (Markovic et al. 2011). White mycelial felts can also develop in shrinkage cracks of the decayed wood (Ryvarden and Gilbertson 1993). After establishment, the perennial basidiome appears relatively rapidly (Mounce 1929, Fig. 1). The infection results in the breakage of treetops, or further infection of the base of the trees and weakening of larger roots, which may lead to eventual windthrow of standing trees.
Hosts – The type species, F. pinicola mostly appears on gymnosperms, such as Abies, Larix, Picea and Pinus, but can also be found on angiosperms such as Acer, Alnus, Betula, Carpinus, Corylus, Elaeagnus, Fagus, Fraxinus, Malus, Populus, Prunus, Pyrus, Quercus, Salix, Sorbus, Tilia, Ulmus (Ryvarden and Gilbertson 1993; Dai 2012). The North American species in the Fomitopsis pinicola species complex have also been reported from Pseudotsuga, Sequioa and Tsuga (Haight et al. 2019). Other Fomitopsis species can be found on Ginkgo, Pinus and various angiosperm genera, such as Betula, Castanopsis, Cinamomum, Citrus, Delonix, Fagus, Eucalyptus Ligustrum, Prunus, Quercus and Tilia (Ryvarden and Gilbertson 1993; Dai 2012; Li et al. 2013; Han et al. 2016; Liu et al. 2019).
Morphological based identification and diversity
Based on morphological evidence, over 40 species were accepted in Fomitopsis (e.g. Ryvarden and Johansen 1980; Gilbertson and Ryvarden 1986; Ryvarden and Gilbertson 1993; Núñez and Ryvarden 2001; Hattori 2001). However, phylogenetic studies showed that the morphologically defined Fomitopsis was polyphyletic and taxa clustered with other brown-rot genera in the antrodia clade (Ortiz-Santana et al. 2013; Han et al. 2016). Han et al. (2016) showed that Pilatoporus and Piptoporus are synonyms of Fomitopsis sensu stricto, while the segregation of Rhodofomes was confirmed and five new genera were proposed. Fomitopsis sensu stricto is characterized by annual to perennial, mostly sessile, occasionally effused-reflexed or substipitate, soft, corky, tough to woody basidiocarps, a dimitic hyphal system with clamped generative hyphae and cylindrical to ellipsoid, hyaline, thin-walled, smooth basidiospores which are negative in Melzer’s reagent, and cause brown rot (Fig. 1).
Fig. 1 Fomitopsis pinicola basidiomes on living European spruce, b causing brown-rot decay on narrow-leafed ash, c–d basidiomes on dead standing conifer tree, e young basidiome on hardwood log, f hyphal structure in the trama, g–h basidiospores. Scale bars: f = 20 µm, g–h = 5 µm.
Molecular based identification and diversity
Comprehensive multigene analyses by Han et al. (2016) accepted ten species in Fomitopsis sensu stricto. Two new Fomitopsis species were described from Brazil, F. flabellata and F. roseoalba (Tibpromma et al. 2017). Fomitopsis flabellata was transferred to Rhodofomitopsis and the new combination Fomitopsis bondartsevae was proposed (Soares et al. 2017). Mating studies and molecular phylogenetic analyses resolved four cryptic lineages in the F. pinicola species complex (Haight et al. 2016), that represents three North American species (F. mounceae, F. ochracea and F. schrenkii), and F. pinicola sensu stricto, which is restricted to Eurasia (Ryvarden and Stokland 2008; Haight et al. 2019). Three new species were proposed by Liu et al. (2019) from Australia (F. eucalypticola), Puerto Rico (F. caribensis), and China (F. ginkgonis).
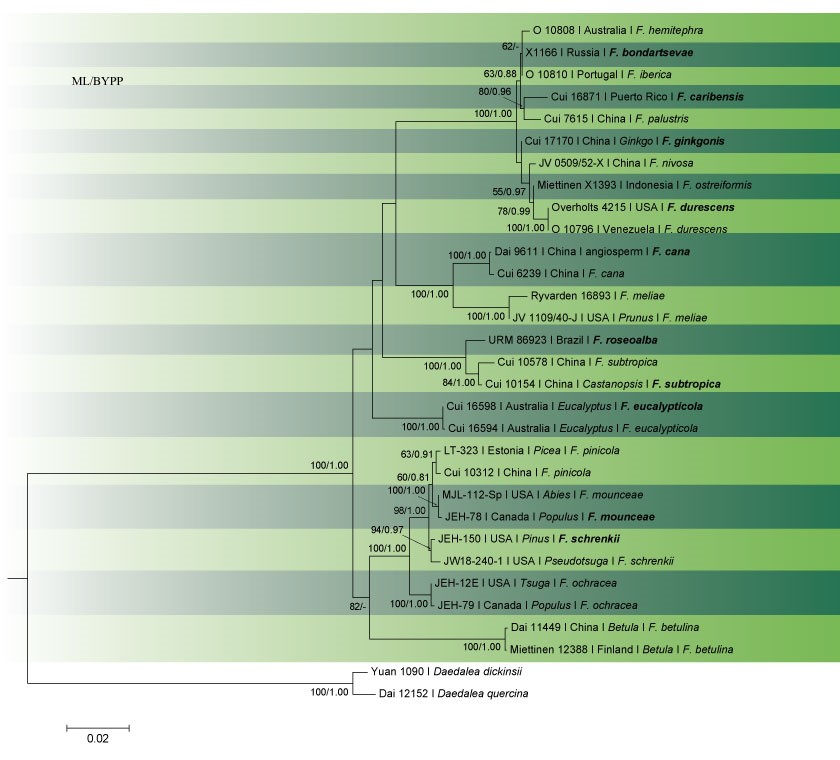
The phylogenetic tree of Fomitopsis presented here is based on analyses of a combined ITS, LSU, tef1 and rpb2 sequence data (Fig 2). In our analyses, it appears that the type of F. bondartsevae is identical to F. iberica and F. hemitephra sensu stricto (Han et al. 2016), which are grouped close to F. palustris and other species formerly discussed in Pilatoporus. Therefore, a thorough revision of the pilatoporus clade is recommended to clarify the status of these species.
Recommended genetic marker (genus level) – LSU
Recommended genetic markers (species level) – ITS, tef1, rpb2
Accepted number of species – There are 104 epithets listed in Index Fungorum (2020). However, only 17 species have DNA sequence data (Table 1).
References – Li et al. (2013) (phylogeny, new species), Han et al. (2016) (phylogeny), Haight et al. 2019 (phylogeny, new species), Floudas et al. (2012) (genome, F. pinicola), Hong et al. (2017) (genome, F. palustris), Liu et al. (2019) (phylogeny, new species).
Table 1 DNA barcodes for accepted species of Fomitopsis. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Strain | ITS | LSU | nSSU | tef1 | rpb2 |
| Fomitopsis betulina | Dai 11449 | KR605798 | KR605737 | KR605895 | KR610726 | KR610816 |
| F. betulina | Miettinen 12388 | JX109856 | JX109856 | – | JX109913 | JX109884 |
| F. bondartsevae | X1166* | JQ700276 | JQ700276 | – | – | – |
| F. cana | Dai 9611* | JX435776 | JX435774 | KR605825 | KR610660 | KR610762 |
| F. cana | Cui 6239 | JX435777 | JX435775 | KR605826 | KR610661 | KR610761 |
| F. caribensis | Cui 16871* | MK852559 | MK860108 | MK860124 | MK900482 | MK900474 |
| F. durescens | Overholts 4215* | KF937293 | KF937295 | KR605835 | – | – |
| F. durescens | O 10796 | KF937292 | KF937294 | KR605834 | KR610669 | KR610766 |
| F. eucalypticola | Cui 16598* | MK852562 | MK860113 | MK860129 | MK900484 | MK900479 |
| F. eucalypticola | Cui 16594 | MK852560 | MK860110 | MK860126 | MK900483 | MK900476 |
| F. ginkgonis | Cui 17170* | MK852563 | MK860114 | MK860130 | MK900485 | MK900480 |
| F. ginkgonis | Cui 17171 | MK852564 | MK860115 | MK860131 | MK900486 | MK900481 |
| F. hemitephra | O 10808 | KR605770 | KR605709 | KR605841 | KR610675 | – |
| F. iberica | O 10810 | KR605771 | KR605710 | KR605842 | KR610676 | KR610771 |
| F. meliae | Ryvarden 16893 | KR605776 | KR605715 | KR605849 | KR610681 | KR610775 |
| F. meliae | JV 1109/40-J | KY264030 | – | – | – | – |
| F. mounceae | JEH-78* | KF169629 | – | – | KF178354 | KF169698 |
| F. mounceae | MJL-112-Sp | KF169636 | – | – | KF178361 | KF169705 |
| F. nivosa | JV 0509/52-X | KR605779 | KR605718 | – | KR610686 | KR610777 |
| F. ochracea | JEH-12E | KF169597 | – | – | KF178322 | KF169666 |
| F. ochracea | JEH-79 | KF169604 | – | – | KF178329 | KF169673 |
| F. ostreiformis | Miettinen X1393 | KC595918 | KC595918 | – | – | – |
| F. palustris | Cui 7615 | KR605780 | KR605719 | – | KR610688 | KR610779 |
| F. pinicola | Cui 10312 | KR605781 | KR605720 | KR605856 | KR610689 | KR610780 |
| F. pinicola | LT-323 | KF169651 | – | – | KF178376 | KF169720 |
| F. roseoalba | URM 86923* | KT189139 | KT189141 | – | – | – |
| F. schrenkii | JEH-150* | KU169365 | – | – | MK236356 | MK208858 |
| F. schrenkii | JW18-240-1 | KF169648 | – | – | KF178373 | KF169717 |
| F. subtropica | Cui 10154* | JQ067652 | JX435772 | – | – | – |
| F. subtropica | Cui 10578 | KR605787 | KR605726 | KR605867 | KR610698 | KR610791 |
Fig. 2 Phylogram generated from RAxML analysis based on combined ITS, LSU, nSSU, tef1 and rpb2 sequence data of Fomitopsis species. Related sequences were obtained from GenBank. Thirty-one strains are included in the analyses, which comprised 4143 characters including gaps. The tree was rooted with Daedalea quercina (Dai 12152) and D. dickinsii (Yuan 1090). Tree topology of the ML analysis was similar to the Bayesian analysis. ML bootstrap values ˃50% and BYPP ˃0.80 are shown respectively near the nodes.


No Comments