18 Sep Plenodomus
Plenodomus Preuss, Linnaea 24:145(1851)
For synonyms see Index Fungorum (2018)
Background
The genus Plenodomus is one of the oldest pleosporalean genera with a long history of taxonomic debate. Preuss (1851) introduced Plenodomus based on P. rabenhorstii (de Gruyter et al. 2013, Ariyawansa et al. 2015a). However, the type material of P. rabenhorstii was lost during World War II (Ariyawansa et al. 2015a) and therefore later Boerema and Van Kesteren (1964) replaced the type species of Plenodomus by P. lingam (sexual morph: Leptosphaeria maculans). Plenodomus species are widely distributed throughout the world with species mainly causing cankers and leaf spots associated with a wide variety of substrates (Wijayawardene et al. 2017b; Farr and Rossman 2018). Plenodomus includes several well known important plant pathogens, such as Pl. biglobosus, P. lindquistii, Pl. tracheiphilus, and P. wasabiae (Marin-Felix et al. 2017). Most of the pathogenic records are for Plenodomus destruens and molecular BLAST results in GenBank show that this species belongs to Valsaceae in Sordariomycetes. Therefore, the pathogenetic virulence of this genus requires further investigation with more taxon sampling and DNA based sequence analyses. There are few sexual records for this genus and recently Tennakoon et al. (2017) introduced Plenodomus sinensis as a sexual morph with an updated molecular phylogeny in this genus.
Classification – Dothideomycetes, Pleosporomycetidae, Pleosporales, Leptosphaeriaceae
Type species – Plenodomus lingam (Tode: Fr.) Höhn., Sber. Akad. Wiss.Wien, Math.-naturw. Kl., Abt. 1 120:463(1911)
Distribution – Worldwide
Disease Symptoms – Foot rot, Dieback, Mal secco of Citrus, wilting
Symptoms may vary according to the host. The typical symptoms consist of red discolouration strands in the xylem of stems, veinal chlorosis, wilt and shedding of leaves resulting in ultimate dieback of twigs and branches (Nachmias et al. 1979; Migheli et al. 2009). In seedbeds, seedlings become yellow, especially the lower leaves resulting in wilt and death of the plant. In the field, plants show a blackening of the tree around the soil level extending upward and downward. The lower part rots, and root system disintegrates. Affected stems may girdle and death of plant will follow (Lopes and Silva 1993).
Hosts – Plenodomus species are recorded from at least 50 plant genera in Apiaceae, Arecaceae, Bignoniaceae, Brassicaceae, Convolvulaceae, Cucurbitaceae, Fabaceae, Gesneriaceae, Lamiaceae, Liliaceae, Moraceae, Oleaceae, Pandanaceae, Poaceae, Ranunculaceae, Rosaceae, Rutaceae, Salicaceae, Santalaceae, Urticaceae and Vitaceae as either saprobes or pathogens (Farr and Rossman 2018).
Morphological based identification and diversity
Plenodomus species are characterized by the ability of their asexual morph to produce scleroplectenchyma in the peridium of the pycnidium, i.e. hyaline cells with thick walls and a relatively small lumen (Boerema et al. 1994). The sexual morph is characterized by papillate, ostiolate ascomata, scleroplectenchymatous cells in the peridium, short pedicellate, cylindrical asci, and cylindrical to ellipsoidal, multi-septate pigmented ascospores (Ariyawansa et al. 2015a). Nevertheless, the simple generic diagnosis such as ‘scleroplectenchyma-producing’ defined by Boerema et al. (1994) which is similar to some species of other genera e.g. Leptosphaeria, has made Plenodomus a large, heterogeneous assemblage and Index Fungorum currently lists 97 epithets. The exact familial placement of these epithets are obscure due to lack of molecular data (<25 taxa have DNA data out of those 97 epithets) and it is necessary to recollect these taxa from type localities, isolate them in axenic culture, and analyse their DNA sequence data to integrate them into appropriate taxonomic ranks. Wijayawardene et al. (2017b) estimated there were 18 species in this genus. Most recently, Marin-Felix et al. (2017) accepted 20 species in their molecular phylogenetic analyses.
Species delimitation in Plenodomus based on morphology is difficult due to the overlapping of morphological characters among many species (de Gruyter et al. 2013). Therefore, DNA sequence data is very important in species identification within this genus.
Molecular based identification and diversity
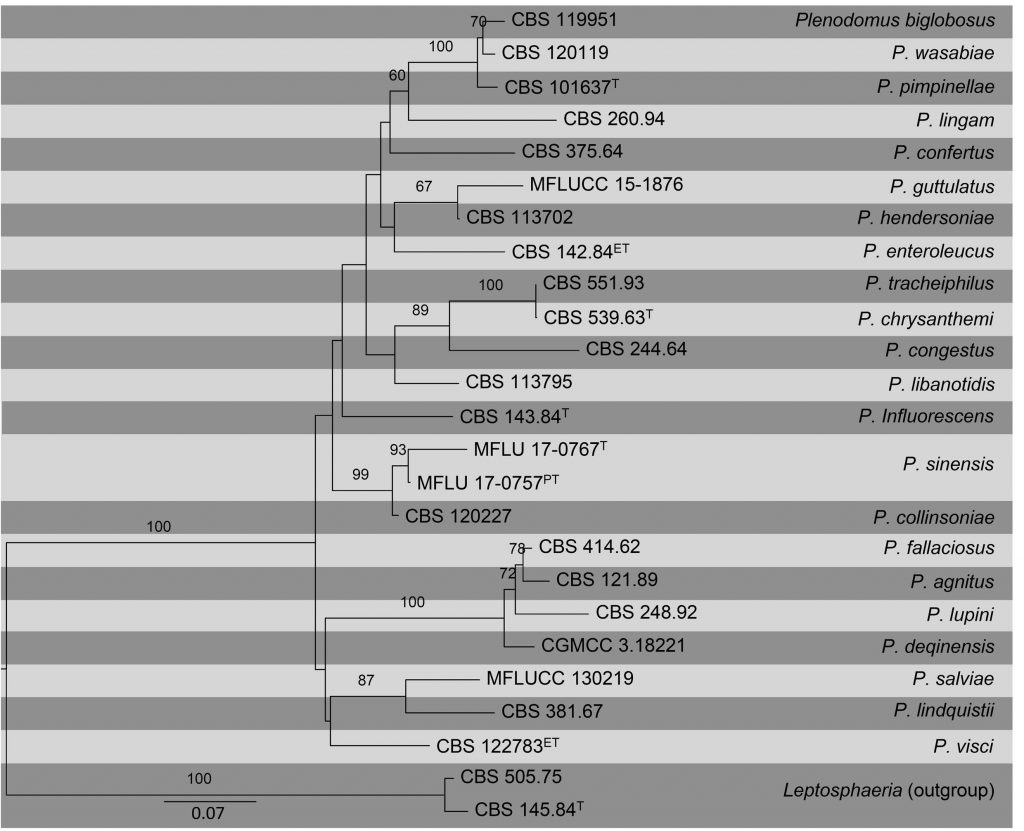
To achieve better generic and species delimitation, phylogenetic studies using ITS, TUB2 and RPB2 were recently performed (Marin-Felix et al. 2017). Phylogenetic studies based on these loci made it possible to reallocate species of Plenodomus to their exact genera (de Gruyter et al. 2013). We update the phylogeny of this genus based on a combined ITS, TUB2 and RPB2 sequence data obtained from available cultures including ex-type, ex-epitype and ex-paratype strains. The topological structure obtained in this study is in accordance with Marin-Felix et al. (2017) and Tennakoon et al. (2017).
Recommended genetic marker (genus level) – LSU
Recommended genetic markers (Species level) – ITS, TUB2 and RPB2
Based on our phylogeny, we observed that TUB2 gives a high resolution compared to other gene regions, such that it can be readily used to determine the placement of Plenodomus species. It is recommended to use a combination of ITS, TUB2 and RPB2 sequence data for a better resolution.
Accepted number of species: There are 97 species epithets in Index Fungorum (2018) under this genus. However, only 22 are accepted.
References: de Gruyter et al. 2013, Ariyawansa et al. 2015a, Marin-Felix et al. 2017, Tennakoon et al. 2017 (morphology, phylogeny).
Table Plenodomus. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate | ITS | RPB2 | TUB2 |
| Leptosphaeria doliolum | CBS 505.75 | JF740205 | KY064035 | JF740144 |
| L. etheridgei | CBS 145.84 | JF740254 | JF740160 | |
| Plenodomus agnitus | CBS 121.89 | JF740194 | KY064036 | KY064053 |
| P. biglobosus | CBS 119951 | JF740198 | KY064037 | KY064054 |
| P. chrysanthemi | CBS 539.63* | JF740253 | KY064038 | KY064055 |
| P. collinsoniae | CBS 120227 | JF740200 | KY064039 | KY064056 |
| P. confertus | CBS 375.64 | AF439459 | KY064040 | KY064057 |
| P. congestus | CBS 244.64 | AF439460 | KY064041 | KY064058 |
| P. deqinensis | CGMCC 3.18221 | KY064027 | KY064034 | KY064052 |
| P. enteroleucus | CBS 142.84* | JF740214 | KY064042 | KT266266 |
| P. fallaciosus | CBS 414.62 | JF740222 | KY064043 | |
| P. guttulatus | MFLUCC 15-1876 | KT454721 | ||
| P. hendersoniae | CBS 113702 | JF740225 | KY064044 | KT266271 |
| P. influorescens | CBS 143.84* | JF740228 | KY064045 | KT266267 |
| P. libanotidis | CBS 113795 | JF740231 | KY064046 | KY064059 |
| P. lindquistii | CBS 381.67 | JF740233 | ||
| P. lingam | CBS 260.94 | JF740235 | KY064047 | KY064060 |
| P. lupine | CBS 248.92 | JF740236 | KY064048 | KY064061 |
| P. pimpinellae | CBS 101637* | JF740240 | KY064062 | |
| P. sinensis | MFLU 17-0767* | MF072721 | ||
| P. sinensis | MFLU 17-0757P* | MF072722 | ||
| P. salvia | MFLUCC 130219 | KT454725 | ||
| P. tracheiphilus | CBS 551.93 | JF740249 | KY064049 | KT266269 |
| P. viscid | CBS 122783* | JF740256 | KY064050 | KY064063 |
| P. wasabiae | CBS 120119 | JF740257 | KT266272 |
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, TUB2 and RPB2 sequence data of Plenodomus species. Related sequences were obtained from GenBank. Twenty-five strains are included in the analyses, which comprise 1695 characters including gaps. Tree was rooted with Leptosphaeria doliolum (CBS 505.75) and L. etheridgei (CBS 145.84). The best scoring RAxML tree with a final likelihood value of -10464.446766 is presented. The matrix had 651 distinct alignment patterns, with 23.12% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.228545, C = 0.263966, G = 0.25175, T = 0.25574; substitution rates AC = 1.629051, AG = 5.903589, AT = 1.764611, CG = 1.424439, CT = 9.08177, GT = 1.000000; gamma distribution shape parameter α = 0.359918. RAxML bootstrap support values ≥60% (BT) are shown respectively near the nodes. The scale bar indicates 0.07 changes. T, ET and PT indicate ex-type, ex-epitype, and ex-paratype strains, respectively.

No Comments