15 Oct Stagonosporopsis
Stagonosporopsis Died. emend. Aveskamp et al., Stud. Mycol. 65: 44. 2010
Stagonosporopsis is a coelomycetous genus in the family Didymellaceae (de Gruyter et al. 2013), accommodating several important phytopathogenic species. Some of the species have described sexual forms in Didymella (Diedicke 1912; Aveskamp et al. 2010). Some Stagonosporopsis species have quarantine importance. Stagonosporopsis andigena is listed in Annex IAI by European Union (EU), meaning its introduction to EU is prohibited. This pathogen is also listed in the A1 list by the European and Mediterranean Plant Protection Organization (EPPO, 2019). Stagonosporopsis chrysanthemi is another species listed in A2 list by EPPO (EPPO, 2019).
Classification – Dothideomycetes, Pleosporomycetidae, Pleosporales, Didymellaceae
Type species – Stagonosporopsis boltshauseri (Sacc.) Died. 1912
Distribution – Worldwide
Disease symptoms – the Black blight of potato, gummy stem blight, ray blight
All plant parts may be attacked by S. chrysanthemi and S. inoxydabilis, however, flowers and cuttings are highly susceptible. Death of flowers and buds, a necrotic lesion on leaves and peduncles of unopened buds, soft rot of cortex of roots and discolouration of bark are the main symptom of the disease. Eventually, plant death occurs (Fox 1998; Pethybridge et al. 2008). In gummy stem blight of cucurbits, symptoms can be observed on all above ground and reproductive parts. Leaf spots are the main diagnostic character of this disease. Most of the circular or triangular shaped spots start at the margin of the leaf or extend towards the margin. The centre of the leaf spot is a lighter shade of brown than the surrounding portion. As leaf spots coalesce leaf blights occur. Actively expanding lesions on leaves, petioles, and pedicels often appear as water-soaked. Cankers may form on crowns, main stems and vines (Keinath 2013). Stagonosporopsis andigena the black blight of potato causal agent affects leaves, petioles and stems causing lesions and premature leaf drop, but does not infect the underground parts. On leaves, the pathogen causes small, blackish concentric lesions. The initial symptoms can be observed on the lower leaves, however, as the disease progresses lesions may also develop in upper leaves as well. Lesions may coalesce and severely affected leaves may turn blackish giving a scorched appearance (French 2001; EFSA panel on plant health 2019).
Hosts – Amaranthaceae, Asteraceae, Campanulaceae, Caryophyllaceae, Cucurbitaceae, Fabaceae, Lamiaceae, Pinaceae, Ranunculaceae, Solanaceae and Valerianaceae.
Morphological based identification and diversity
Stagonosporopsis is characterized by ellipsoidal to subglobose, aseptate to 3 septate conidia and sexual morph with ellipsoidal, fusiform or obovoid, 1 septate ascospore (Aveskamp et al. 2010; Chen et al. 2015). Stagonosporopsis was originally separated from Ascochyta by Diedicke (1912) based on the occasional formation of multi-septate (Stagonospora-like) conidia. In the phylogenetic reassessment of Didymellaceae (Aveskamp et al. 2010) based on the sequences LSU and ITS of the nrDNA and TUB2 region, multiple Phoma species, including P. ligulicola, were recovered in a high supported clade with the interpretive types of the genus Stagonosporopsis; S. actaeae (Boerema et al. 1997, 2004). In addition, S. tanaceti shows morphological similarity to S. inoxydabilis but can be differentiated by the faster growth rate, larger conidia, presence of chlamydospores, and lack of ascomata in culture (Vaghefi et al. 2012). Morphological characters overlap between the species in this genus and species are primarily separated based on molecular data.
Molecular based identification and diversity
Most comprehensive multigene phylogeny analyses for this genus were performed by Aveskamp et al. (2010), Vaghefi et al. (2012), Hyde et al. (2014), Chen et al. (2015, 2017) and Jayasiri et al. (2019). Five-marker phylogeny of the Stagonosporopsis spp. for which these DNA sequence data are available is shown (Table ).
Identification of Stagonosporopsis species associated with ray blight of Asteraceae can be achieved through multi-locus sequence typing (Aveskamp et al. 2010; Vaghefi et al. 2012) and also with a species-specific multiplex PCR assay developed by Vaghefi et al. (2016). This assay is based on a four-primer PCR that targets the intergenic spacer of the nrDNA of the ray blight pathogens, producing species-specific amplicons of ~ 560 in S. chrysanthemi, ~ 630 bp in S. inoxydabilis and ~ 400 bp in S. tanaceti, which can be easily differentiated on an agarose gel (Vaghefi et al. 2016).
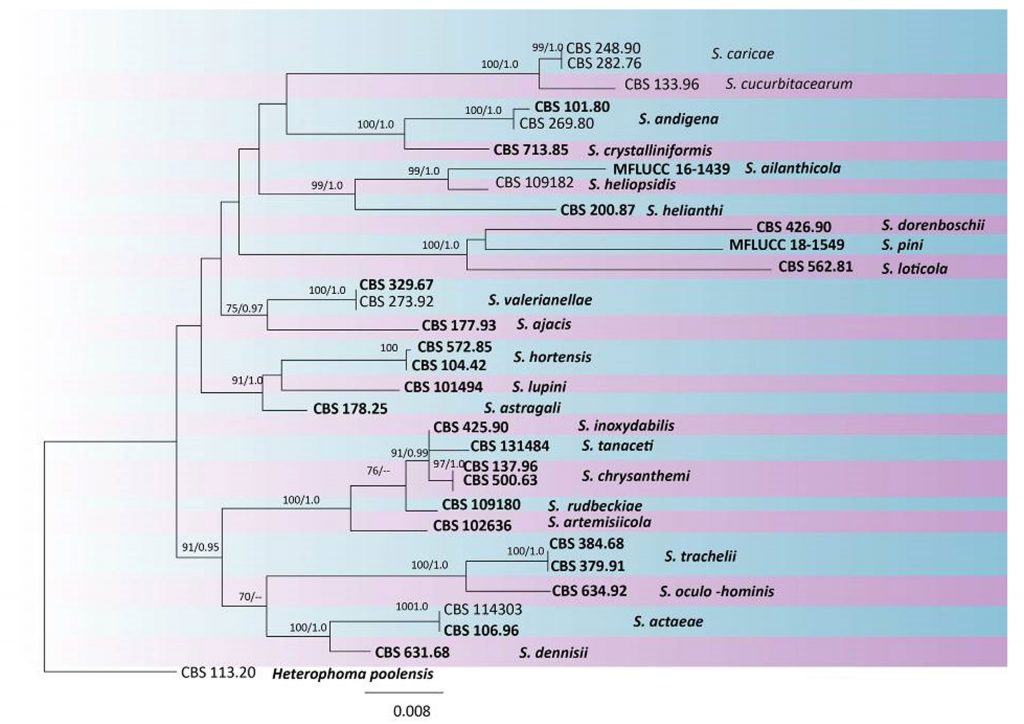
This study reconstructs the phylogeny of Stagonosporopsis based on analyses of a combined ITS, LSU, RPB2 and TUB2 sequence data (Fig ). The phylogenetic tree is updated with recently introduced Stagonosporopsis species and corresponds to previous studies (Chen et al. 2017, Jayasiri et al. 2019).
Recommended genetic markers (genus level) – ITS
Recommended genetic markers (species level) – TUB2 and RPB2
Accepted number of species: There are 55 species in Index Fungorum (2019) and only 24 species are accepted/ have molecular data in this genus.
References: Chen et al. 2015, 2017, Jayasiri et al. 2019 (morphology, phylogeny).
Table Details of the Stagonosporopsis isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher stains are in bold.
| Species | Culture | GenBank Accession numbers | |||
| LSU | ITS | RPB2 | TUB2 | ||
| Stagonosporopsis actaeae | CBS 106.96* | GU238166 | GU237734 | KT389672 | GU237671 |
| CBS 114303 | KT389760 | KT389544 | – | KT389847 | |
| S. ailanthicola | MFLUCC 16-1439* | KY100874 | KY100872 | KY100876 | KY100878 |
| S. ajacis | CBS 177.93* | GU238168 | GU237791 | KT389673 | GU237673 |
| S. andigena | CBS 269.80 | GU238170 | GU237817 | – | GU237675 |
| CBS 101.80 | GU238169 | GU237714 | – | GU237674 | |
| S. artemisiicola | CBS 102636 | GU238171 | GU237728 | KT389674 | GU237676 |
| S. astragali | CBS 178.25 | GU238172 | GU237792 | – | GU237677 |
| S. caricae | CBS 248.90 | GU238175 | GU237807 | – | GU237680 |
| CBS 282.76 | GU238177 | GU237821 | – | GU237682 | |
| S. chrysanthemi | CBS 500.63 | GU238190 | GU237871 | – | GU237695 |
| CBS 137.96 | GU238191 | GU237783 | – | GU237696 | |
| S. crystalliniformis | CBS 713.85* | GU238178 | GU237903 | KT389675 | GU237683 |
| S. cucurbitacearum | CBS 133.96 | GU238181 | GU237780 | KT389676 | GU237686 |
| S. dennisii | CBS 631.68* | GU238182 | GU237899 | KT389677 | GU237687 |
| S. dorenboschii | CBS 426.90* | GU238185 | GU237862 | KT389678 | GU237690 |
| S. helianthi | CBS 200.87* | KT389761 | KT389545 | KT389683 | KT389848 |
| S. heliopsidis | CBS 109182 | GU238186 | GU237747 | KT389679 | GU237691 |
| S. hortensis | CBS 104.42 | GU238198 | GU237730 | KT389680 | GU237703 |
| CBS 572.85 | GU238199 | GU237893 | KT389681 | GU237704 | |
| S. inoxydabilis | CBS 425.90* | GU238188 | GU237861 | KT389682 | GU237693 |
| S. loticola | CBS 562.81* | GU238192 | GU237890 | KT389684 | GU237697 |
| S. lupini | CBS 101494* | GU238194 | GU237724 | KT389685 | GU237699 |
| S. oculo –hominis | CBS 634.92* | GU238196 | GU237901 | KT389686 | GU237701 |
| S. pini | MFLUCC 18-1549* | MK348019 | MK347800 | MK434860 | – |
| S. rudbeckiae | CBS 109180 | GU238197 | GU237745 | – | GU237702 |
| S. tanaceti | CBS 131484* | JQ897461 | NR_111724- | JQ897496 | |
| S. trachelii | CBS 379.71 | GU238173 | GU237850 | KT389687 | GU237678 |
| CBS 384.68 | GU238174 | GU237856 | – | GU237679 | |
| S. valerianellae | CBS 273.92 | GU238200 | GU237819 | – | GU237705 |
| CBS 329.67* | GU238201 | GU237832 | GU237706 | ||
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU, RPB2 and TUB2 sequence data of Stagonosporopsis species. Related sequences were obtained from GenBank. Thirty-two are included in the analyses, which comprise 2756 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Heterophoma poolensis (CBS 113.20). Tree topology of the ML analysis was similar to the MP. The best scoring RAxML tree with a final likelihood value of -8386.622374 is presented. The matrix had 388 distinct alignment patterns, with 11.83% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.245798, C = 0.237676, G = 0.273178, T = 0.243348; substitution rates AC = 1.882600, AG = 4.123164, AT = 2.015920, CG = 0.907833, CT = 12.626910, GT = 1.000000; gamma distribution shape parameter α = 1.105097. RAxML bootstrap support values ≥50% and Bayesian posterior probabilities ≥0.95 (BYPP) are shown respectively near the nodes. Ex-type strains are in bold.

No Comments