26 Oct Golovinomyces
Golovinomyces (U. Braun) V.P. Heluta, Biol. Zh. Armenii 41: 357 (1988)
Background
Braun (1978) introduced Golovinomyces as a section of Erysiphe sensu lato and Heluta (1988a) raised it to genus rank. Braun (1999) and Braun and Takamatsu (2000) accepted Golovinomyces as a distinct genus and established a new tribe, Golovinomyceteae. This is a strictly herb-parasitic genus in the Erysiphaceae. Host-parasite co-speciation was reported between Golovinomyces and Asteraceae hosts using molecular phylogenetic analyses (Matsuda and Takamatsu 2003). It was suggested that Golovinomyces first acquired parasitism on Asteraceae and then diverged to the host tribes Astereae, Cardueae, Heliantheae and Lactuceae. Bremer (1994) pointed out that Golovinomyces may have originated in South America and the geographic distribution expanded into the Northern Hemisphere. However, Takamatsu et al. (2006) suggest that Golovinomyces originated in the Northern Hemisphere, and not in South America. Fabro et al. (2008) profiled genome-wide expression on haustorium formation of G. cichoracearum in Arabidopsis. Research to understand pathogenesis towards plants has been undertaken. A draft whole genome of G. magnicellulatus, the causal agent of phlox powdery mildew was provided by Farinas et al. (2019). McKernan et al. (2020) identified 82 genes associated with resistance to G. chicoracearum, the causal agent of powdery mildew in cannabis.
Classification – Ascomycota, Pezizomycotina, Leotiomycetes, Leotiomycetidae, Erysiphales, Erysiphaceae
Type species – Golovinomyces cichoracearum (DC.) V.P. Heluta
Distribution – Worldwide (Mainly in northern hemisphere)
Disease symptoms – powdery mildew
Hosts – Has a wide range of hosts including Asteraceae, Boraginaceae, Cucurbitaceae, Malvaceae, Fabaceae, Lamiaceae, Polygonaceae, Scrophulariaceae, Solanaceae and Verbenaceae.
Pathogen biology, disease cycle and epidemiology
Discussed under Erysiphaceae.
Morphological based identification and diversity
Golovinomyces is characterized by chasmothecia with mycelioid appendages, several, mostly 2-spored asci, an asexual morph with catenescent conidia that lack fibrosin bodies, and mostly nipple-shaped appressoria (Braun 1978; Qiu et al. 2020a). Heluta (1988a) reallocated Erysiphae cichoracearum to Golovinomyces and now nearly all species of E. cichoracearum are assigned to Golovinomyces. Braun (1987) confined E. cichoracearum to powdery mildews on hosts of Asteraceae and assigned specimens on hosts belonging to other plant families to Erysiphe orontii. Braun and Cook (2012) split G. cichoracearum into several species based on molecular analyses of this complex which suggested a co-evolutionary relationship between Golovinomyces species and tribes of Asteraceae (Matsuda and Takamatsu 2003). Golovinomyces cynoglossi sensu lato, a complex of morphologically similar powdery mildews on the plant family Boraginaceae, was reassessed by Braun et al. (2018) and split into G. asperifoliorum, G. asperifolii and G. cynoglossi based on sequence analyses, biological aspects and morphological differences. Braun et al. (2019) revisited G. orontii and Qiu et al. (2020b) epitypfied and confirmed Erysiphe cucurbitacearum was a synonym of G. tabaci.
Molecular based identification and diversity
A comprehensive phylogenetic analysis by Takamatsu et al. (2013) resulted in a polyphyletic complex that split into three genetically distinct clades. Golovinomyces ambrosiae and G. spadiceus were considered as separate species by Braun and Cook (2012). However, phylogenetic analyses of ITS and 28S rDNA sequences by Takamatsu et al. (2013), including Golovinomyces species on Asteraceae, found that these two species that occur on Asian species of Eupatorium and a multitude of other hosts, including those on other plant families, formed a single large, unresolved clade (lineage III in Takamatsu et al. (2013)). The taxonomic interpretation posed a serious problem as G. ambrosiae and G. spadiceus were treated as two morphologically differentiated species. Hence, the resolution based only on ITS sequence data was considered insufficient to distinguish closely allied species. Most subsequent authors followed the taxonomic treatments in Braun and Cook (2012) and recognized G. ambrosiae and G. spadiceus as separate species, within lineage III, based on morphological differences (Qiu et al. 2020a). However, there is minimal multi loci data for the powdery mildews currently available. Most of the research involves the intra-specific genetic diversity in species such as Blumeria graminis (Walker et al. 2011), Erysiphe necator (Oliveira et al. 2015), Golovinomyces orontii (Pirondi et al. 2015a) and Podosphaera xanthii (Pirondi et al. 2015b). Based on ITS and D1/D2 domain of 28S sequence data, Braun et al. (2019) introduced G. bolayi and G. vincae. Nayak and Bandamaravuri (2019) developed species-specific PCR primers CgF2 and CgR2 for G. orontii (the causal agent of powdery mildew in cucurbits), based on partial ITS and 5.8S rDNA, which resulted in a 233bp fragment of G. orontii.
Recommended genetic markers (genus level) – ITS, LSU
Recommended genetic markers (species level) – Comprehensive applications of multi loci approaches to solve complex taxonomic-phylogenetic problems connected with the species level classification of the powdery mildews are lacking. The phylogenetic analyses of multi loci sequence data, including ITS and LSU, IGS, tub2, chs, and consideration of morphological characters resolve species delimitation in a heterogeneous complex within Golovinomyces.
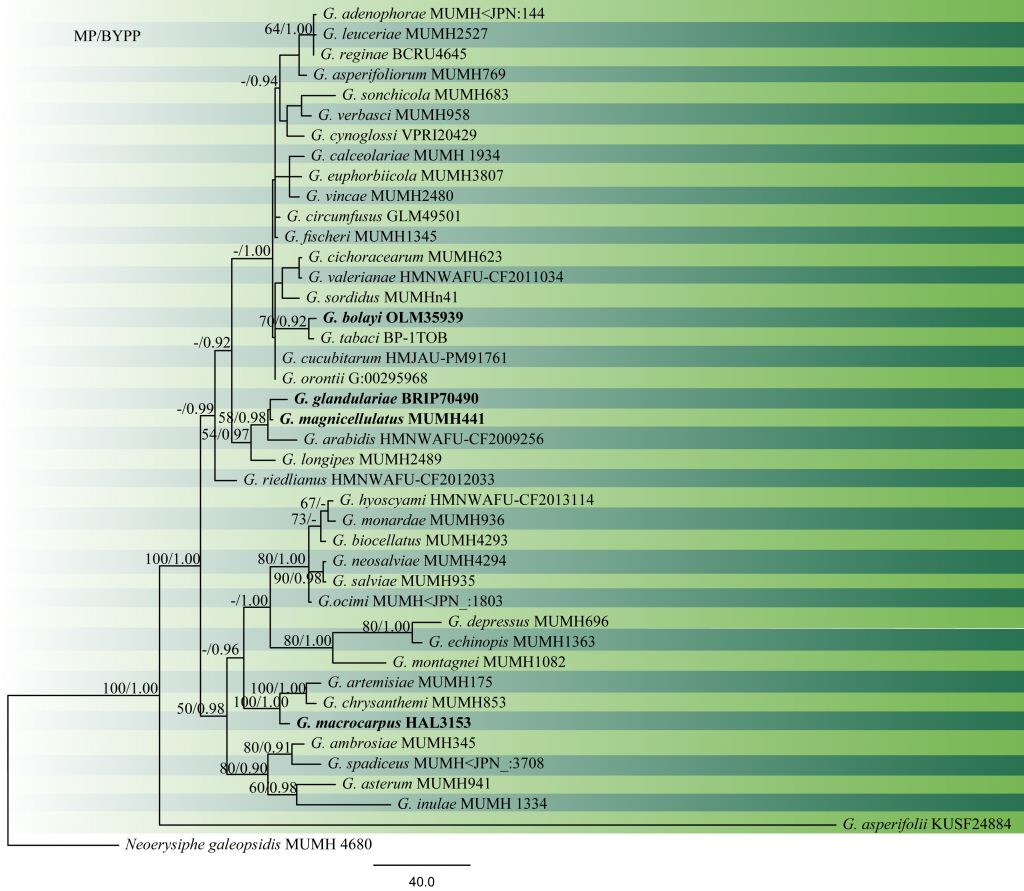
Accepted number of species – There are 81 epithets listed in Index Fungorum (2020), however, only 41 have molecular data (Table 1, Fig. 1).
References – Braun 1978, 1987, Heluta 1988 (morphology); Braun and Cook 2012, Takamatsu et al. 2013, Braun et al. 2019, Qiu et al. 2020a,b (morphology and phylogeny).
Table 1 DNA barcodes for accepted species of Golovinomyces. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species | Strain no | ITS | LSU |
| Golovinomyces adenophorae | MUMH<JPN_144 | LC516963 | AB077632 |
| G. ambrosiae# | MUMH345 | AB077642 | AB077641 |
| G. arabidis | HMNWAFU-CF2009256 | KR048081 | KR048149 |
| G. artemisiae# | MUMH175 | AB077637 | AB077636 |
| G. asperifolii | KUS-F24884 | MH189697 | MH189696 |
| G. asperifoliorum | MUMH769 | AB077684 | AB077684 |
| G. asterum# | MUMH941 | AB769417 | AB769418 |
| G. biocellatus# | MUMH4293 | AB307671 | AB307671 |
| G. bolayi# | OLM 35939* | LC417106 | – |
| G. calceolariae | MUMH 1934 | AB430810 | AB430810 |
| G. chrysanthemi | MUMH853 | AB077654 | AB077653 |
| G. cichoracearum# | MUMH623 | AB077660 | AB077660 |
| G. circumfusus# | GLM49501 | MK452630 | MK452703 |
| G. cucubitarum | HMJAU-PM91761 | MK937796 | MK937801 |
| G. cynoglossi# | VPRI20429 | AB769455 | – |
| G. depressus | MUMH696 | AB077675 | AB077676 |
| G. echinopis | MUMH1363 | AB769414 | – |
| G. euphorbiicola | MUMH3807 | AB769460 | – |
| G. fischeri | MUMH1345 | AB769451 | AB769452 |
| G. glandulariae | BRIP 70490* | NR_166303 | MN539541 |
| G. hyoscyami | HMNWAFU-CF2013114 | KR048155 | KR048155 |
| G. inulae | MUMH1334 | AB769428 | – |
| G. leuceriae | MUMH2527 | AB246766 | – |
| G. longipes# | MUMH2489 | AB769440 | – |
| G. macrocarpus | HAL 3153* | NR_154105 | – |
| G. magnicellulatus# | MUMH441* | AB077647 | AB077646 |
| G. monardae# | MUMH936 | AB307668 | AB077691 |
| G. montagnei | MUMH1082 | AB769413 | – |
| G. neosalviae# | MUMH4294 | AB307673 | – |
| G. ocimi | MUMH<JPN_:1803 | LC306656 | – |
| G. orontii# | G:00295968 | LC417099 | – |
| G. riedlianus | HMNWAFU-CF2012033 | KR048088 | KR048157 |
| G. reginae | BCRU4645 | AB246759 | – |
| G. salviae | MUMH935 | AB769437 | AB077690 |
| G. sonchicola# | MUMH683 | AB077673 | AB077672 |
| G. sordidus# | MUMHn41 | AB077658 | AB077657 |
| G. spadiceus# | MUMH<JPN_:3708 | LC306664 | – |
| G. tabaci | BP-1TOB | AF229013 | AB022412 |
| G. valerianae | HMNWAFU-CF2011034 | KR048090 | KR048159 |
| G. verbasci# | MUMH958 | AB769468 | AB769469 |
| G. vincae | MUMH2480 | AB769444 | – |
Fig 1 Phylogram generated from MP analysis based on combined sequences of ITS and LSU sequences of all species of Golovinomyces with molecular data. Related sequences were obtained from GenBank. Fourty-two taxa are included in the analyses, which comprise 1401characters including gaps, of which 848 characters are constant, 392 characters are parsimony-uninformative and 161 characters parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of ten equally most parsimonious trees with a length of 927 steps (CI = 0.740, RI=0.699, RC = 0.517, HI = 0.260) in the second tree. The tree was rooted with Neoerysiphe galeopsidis (MUMH 4680). MP bootstrap support value ≥50% and BYPP ≥0.9 are shown respectively near the nodes. Ex-type strains are in bold.

No Comments