26 Apr Coniella
Coniella Höhn., Ber. dt. bot. Ges. 36(7): 316 (1918)
= Pilidiella Petr. & Syd., Beih. Reprium nov. Spec. Regni veg. 42(1): 462 (1927) [1926]; = Schizoparme Shear, Mycologia 15(3): 120 (1923)
For more synonyms see Index Fungorum (2018)
Background
Coniella Höhn. is a cosmopolitan genus which was introduced by von Höhnel (1918) and is typified by Coniella pulchella Höhn. (= Coniella fragariae (Oudem.) B. Sutton). Many Coniella species are known as plant pathogens causing foliar, fruit, leaf, stem and root diseases on a wide range of hosts, including some economically important hosts and have gained considerable attention from the phytopathological community (van Niekerk et al. 2004; Alvarez et al. 2016; Chethana et al. 2017). Several species in this genus have a saprobic lifestyle, occurring in leaf litter, rotting bark and in soil (Alvarez et al. 2016). Several species also occur as endophytes (Alvarez et al. 2016), parasites on unrelated hosts (C. straminea Samuels et al. 1993), and as secondary invaders of plants tissues infected by other organisms or injured by other causes (Ferreira et al. 1997).
Classification– Sordariomycetes, Sordariomycetidae, Diaporthales, Schizoparmaceae
Type species – Coniella fragariae (Oudem.) B. Sutton, Mycol. Pap. 141: 47 (1977)
Distribution – Worldwide
Disease Symptoms – foliar, fruit, stem and root lesions, white rot, crown rot.
On leaves, lesions are marginal, irregular with various shaded brown centres. Light brown specks gradually become reddish-brown larger specks causing wilting and dieback. Fruits may shrivel and change colour to brown.
Diseases on major economic hosts including white rot on grapes (Coniella diplodiella and Coniella vitis; Chethana et al. 2017), fruit and leaf diseases of strawberry (C. castaneicola; Mass 1998), cankers, crown rots, die backs, fruit rots, leaf spots, shoot blights, and twig blights on pomegranates (C. granati; Mirabolfathy et al. 2012; Chen et al. 2014).
Hosts – Wide variety of hosts belonging to Combretaceae, Malvaseae, Myrtaceae, Rosaceae and Vitaceae. Some Coniella species exhibit high host specificity (C. destruens and C. eucalyptorum on Eucalyptus, C. quercicola on Quercus sp., C. crousii on Terminalia sp., C. diplodiella and C. diplodiopsis on Vitis sp., and C. tibouchinae on Tibouchina sp.; Alvarez et al. 2016).
Morphological based identification and diversity
Coniella has been subjected to comprehensive morpho-molecular studies and has undergone several taxonomic refinements over the years (Sutton 1980; Nag Raj 1993; Rossman et al. 2007; van Niekerk et al. 2004; Alvarez et al. 2016). Sutton (1980), Nag Raj (1993) regarded Pilidiella as a synonym of Coniella based on conidial morphology. Samuels et al. (1993) stated Schizoparme as the sexual morph and positioned it in Melanconidaceae. Later, Castlebury et al. (2002) named Pilidiella and Coniella as Schizoparme complex and showed their distinct lineage in Diaporthales. Following Castlebury et al. (2002), Rossman et al. (2007) established a new family, Schizoparmaceae, including the above three genera viz. Coniella, Pilidiella and Schizoparme. Maharachchikumbura et al. (2015, 2016) and Wijayawardene et al. (2016, 2018) accepted Schizoparmaceae as a well-established family comprising Coniella, Pilidiella and Schizoparme. Alvarez et al. (2016) showed that Coniella, Pilidiella and Schizoparme formed a monophyletic clade in Schizoparmaceae, and proposed to adopt Coniella (the older asexual typified name) over Pilidiella and Schizoparme agreeing with Art. 59.1 of the International code of nomenclature for algae, fungi and plants.
Based on conidial pigmentation, van der Aa (in von Arx 1973) and von Arx (1981) treated Coniella and Pilidiella as separate genera, Coniella having dark brown conidia and Pilidiella having hyaline conidia. However, Sutton (1980) and Nag Raj (1993) rejected conidial pigmentation as a distinguishing character and synonymized Pilidiella under the older name, Coniella. Since the introduction of molecular data in species delimitation, many studies have demonstrated that these two asexual genera should be distinct (Castlebury et al. 2002; van Niekerk et al. 2004; Wijayawardene et al. 2016). Due to the many species complexes and similar morphological characters, Alvarez et al. (2016) stated that new species of Coniella must be identified based on both DNA sequence data and morphological characters. Following Alvarez et al. (2016), Chethana et al. (2017) adapted a morphological approach in conjunction with multi-gene phylogeny and Genealogical Concordance Phylogenetic Species Recognition (GCPSR) approach in defining species boundaries.
Colony and conidial morphology are the primary characters to identify species within this genus (Ellis 1971, 1976; Simmons 1992). In terms of morphological characters, Coniella species share several characteristics including conidiomatal anatomy, conidiophores and conidiogenesis. However, morphological characters such as conidial colour, shape, size, presence of basal or lateral mucoid appendages, germ slits, guttules, and cultural characteristics differ depending on the species.
Molecular based identification and diversity
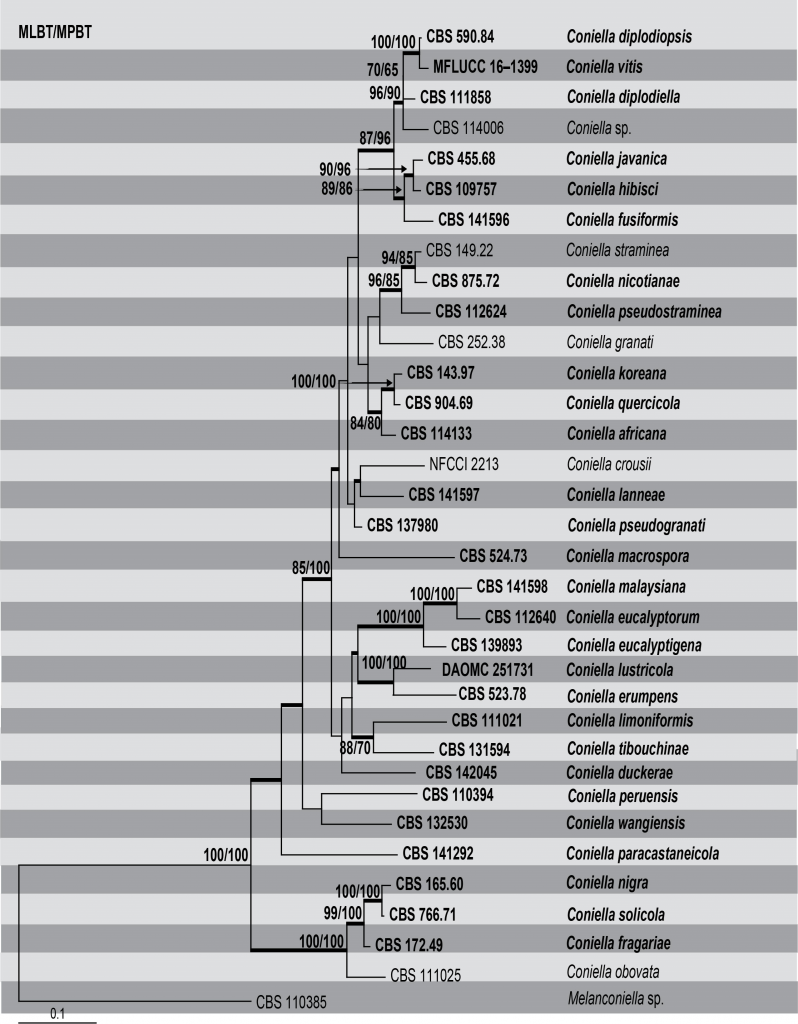
Although Coniella has received much attention, few phylogenetic studies have been conducted. Castlebury et al. (2002) first determined the phylogeny of Coniella in Diaporthales using the large subunit (LSU) nuclear ribosomal DNA (nrDNA) sequence data. Following Castlebury et al. (2002), most studies have used single-gene phylogeny in resolving Coniella. Van Niekerk et al. (2004) used four genes (LSU, ITS, TEF1-α and His3), whereas Miranda et al. (2012) used two genes (ITS and LSU) in their single gene phylogenies. Wijayawardene et al. (2016) combined the latter genes in their phylogenetic analysis. Confusion and inconsistencies revealed by Wijayawardene et al. (2016) leading to poor species delimitation in Coniella were addressed by Alvarez et al. (2016) using a multi-gene phylogenetic approach (ITS, LSU, RPB2 and TEF1-α). Chethana et al. (2017) resolved the taxonomy by combining multi-gene phylogenetic analysis together with GCPSR (ITS, LSU, His3 and TEF1-α). In this section, we reconstruct the phylogeny (Table, Fig) of Coniella based on a combined ITS, LSU, His3 and TEF1-α sequence data. Phylogeny generated herein depicts 34 well-supported clades corresponding to 34 species. The phylogenetic analysis is similar to that of Alvarez et al. (2016) and Chethana et al. (2017). However, the current study includes several new taxa which were introduced recently. Since this genus is of importance to plant pathology, a descriptive study of these species, especially their population dynamics, comparative and functional genomics, will contribute to understanding the pathogenic potential and ecological roles of Coniella species infecting agricultural crops.
Recommended genetic markers (genus level) – LSU and ITS
Recommended genetic markers (species level) – ITS, LSU, TEF 1-α, RPB2 and His3
For the preliminary identification of Coniella species, LSU and ITS gene regions are recommended (Castlebury et al. 2002; van Niekerk et al. 2004; Wijayawardene et al. 2016). Combined analysis using ITS, LSU, TEF1-α, RPB2 and His3 (selection of 4 genes) should be used in resolving species, with recommended primers (Alvarez et al. 2016; Chethana et al. 2017).
Accepted number of species: There are 60 species epithets in Index Fungorum (2018) under this genus. However, only 34 are accepted.
References: van Niekerk et al. 2004, Maharachchikumbura et al. 2015, 2016, Alvarez et al. 2016 (morphology and phylogeny), Chethana et al. 2017 (morphology, phylogeny and pathogenicity).
Table Coniella. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate | ITS | His3 | LSU | TEF 1-a |
| C. africana | CBS 114133* | AY339344 | AY339309 | AY339293 | AY339364 |
| C. crousii | NFCCI 2213 | HQ264189 | – | – | – |
| C. diplodiella | CBS 111858* | AY339323 | AY339297 | AY339284 | AY339355 |
| C. diplodiopsis | CBS 590. 84* | AY339334 | AY339308 | AY339288 | AY339359 |
| C. duckerae | CBS 142045* | KY924929 | – | – | – |
| C. erumpens | CBS 523.78* | KX833533 | – | KX833361 | KX833630 |
| C. eucalyptigena | CBS 139893* | KR476725 | – | KR476760 | – |
| C. eucalyptorum | CBS 112640* | AY339338 | – | AY339290 | KX833637 |
| C. fragariae | CBS 172.49* | AY339317 | – | AY339282 | AY339352 |
| C. fusiformis | CBS 141596* | KX833576 | – | KX833397 | KX833674 |
| C. granati | CBS 252.38 | AY339342 | – | AY339291 | AY339362 |
| C. hibisci | CBS 109757 | KX833581 | – | – | KX833689 |
| C. javanica | CBS 455.68* | KX833583 | – | KX833403 | KX833683 |
| C. koreana | CBS 143.97* | KX833584 | – | AF408378 | KX833684 |
| C. lanneae | CBS 141597* | KX833585 | – | KX833404 | KX833685 |
| C. limoniformis | CBS 111021* | AY339346 | AY339310 | KX833405 | KX833686 |
| C. lustricola | DAOMC 251731* | MF631778 | – | MF631799 | MF651899 |
| C. macrospora | CBS 524.73* | AY339343 | – | AY339292 | AY339363 |
| C. malaysiana | CBS 141598* | KX833588 | KX833406 | KX833688 | |
| C. hibisci | CBS 109757* | KX833589 | – | AF408337 | KX833689 |
| C. nicotianae | CBS 875.72* | KX833590 | – | KX833407 | KX833690 |
| C. nigra | CBS 165.60* | AY339319 | – | KX833408 | KX833691 |
| C. obovata | CBS 111025 | AY339313 | – | KX833409 | KX833692 |
| C. paracastaneicola | CBS 141292* | KX833591 | – | KX833410 | KX833693 |
| C. peruensis | CBS 110394* | KJ710463 | – | KJ710441 | KX833695 |
| C. pseudogranati | CBS 137980* | KJ869132 | – | KJ869189 | – |
| C. pseudostraminea | CBS 112624* | KX833593 | – | KX833412 | KX833696 |
| C. quercicola | CBS 904.69* | KX833595 | – | KX833414 | KX833698 |
| C. solicola | CBS 766.71* | KX833597 | – | KX833416 | KX833701 |
| Coniella sp. | CBS 114006 | AY339347 | AY339311 | AY339295 | KX833703 |
| C. straminea | CBS 149.22 | AY339348 | AY339312 | AY339296 | AY339366 |
| C. tibouchinae | CBS 131594* | JQ281774 | – | JQ281776 | JQ281778 |
| C. vitis | MFLUCC 16–1399* | KX890008 | KX890033 | KX890083 | KX890058 |
| C. wangiensis | CBS 132530* | NR-111764 | – | NG-042686 | KX833705 |
| Melanconiella sp. | CBS 110385 | KX833599 | – | KX833420 | KX833707 |
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU, Histone and TEF1-a sequence data of Coniella species. Related sequences were obtained from GenBank. Thirty four strains are included in the analyses, which comprise 2877 characters including gaps. The tree was rooted with Melanconiella sp. (CBS 110385). Tree topology of the ML analysis was similar to the ones generated from MP and BI (Figures not shown). The best scoring RAxML tree with a final likelihood value of -14885.549943 is presented. The maximum parsimonious dataset consisted of constant characters 2116, 509 parsimony-informative and with 23.84% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.249654, C = 0.245318, G = 0.256486, T = 0.248542; substitution rates AC = 0.977807, AG = 2.195640, AT = 1.226082, CG = 0.712360, CT = 4.190875, GT = 1.000000; gamma distribution shape parameter α = 0.137391. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 2459 steps (CI = 0.525, RI = 0.571, RC = 0.300, HI = 0.475) in the first tree. RAxML and maximum parsimony bootstrap support value ≥ 50% (BT) are shown respectively near the nodes. Bayesian posterior probabilities ≥ 0.95 (PP) and indicated as thickened black branches. The scale bar indicates 0.1 changes. The ex-type strains are in bold.

No Comments