17 Sep Elsinoe
Elsinoe Racib. [as ‘Elsinoë‘], Parasit. Alg. Pilze Java’s (Jakarta) 1: 14 (1900)
Background
Elsinoe was introduced by Raciborski (1900) based on E. canavaliae (Hyde et al. 2013; Jayawardena et al. 2014). von Arx & Müller (1975) placed this genus in Myriangiaceae based on the nature of its pseudoascostromata and parasitic nature. Later, the genus was placed in family Elsinoaceae (Barr 1979; Kirk et al. 2001; Lumbsch and Huhndorf 2007, 2010; Hyde et al. 2013; Jayawardena et al. 2014; Wijayawardene et al. 2017b). Elsinoe is characterized by forming scab-like lesions with pseudoascostromata containing three to eight bitunicate asci in each locule (Jayawardena et al. 2014).The asexual morph is the acervular coelomycetous Sphaceloma de Bary (Wijayawardene et al. 2012, 2017a). Jenkins (1932) proposed a connection between Sphaceloma and Elsinoe. As the sexual morph is not common in nature, morphological based identification of Elsinoe species is difficult. The asexual morph Sphaceloma frequently occurs in nature, however, its morphological characters overlap making identification of the species difficult. Examination of specimens collected in the field is also problematic due to the lack of fertile structures. Isolation of Elsinoë species is also challenging due to their slow growth (Jenkins 1932). In past, scab symptoms have been considered as a major character in recognizing the presence of fungi belonging to this genus, when sporulation is absent (Bitancourt and Jenkins 1949). Fan et al. (2017) suggested that even if spores are absent, species can be named if they have the support of successful isolations, resulting in cultures having common characteristics of the genus. The colonies of this genus are slow growing, raised, cerebriform or corrugated, dark red, orange or brown. If cultures cannot be obtained they should be considered as doubtful species until fertile specimens or pure cultures are obtained (Fan et al. 2017).
Many studies over the past decade have identified secondary metabolites of this genus (Hyde et al. 2013). Elsinochrome is a non-host selective, light-activated polyketide-derived toxin produced by Elsinoe species (Chung and Liao 2008). This is a red-pigmented secondary metabolite mainly produced by E. fawecettii (Yang and Chung 2010). Elsinopirini is a decalin polyketide isolated as a colourless oil from E. pyri (Surup et al. 2018). Production of these secondary metabolites can be used in chemotaxonomy.
Classification – Dothideomycetes, Dothideomycetidae, Myriangiales, Elsinoaceae
Type species – Elsinoe canavaliae Racib. [as ‘canavalliae’], Parasit. Alg. Pilze Java’s (Jakarta) 1: 14 (1900)
Distribution – Worldwide
Disease Symptoms – Scab, Anthracnose of grapevine
Many species cause scab like blemishes (Jayawardena et al. 2014). They can affect leaves, stems and fruits affecting the appearance as well as reducing the yield. Infected organs of some hosts (Cassava) develop severe distortions (Guatimosim et al. 2015).
Hosts – All members of this genus are specialized plant pathogens causing diseases on many economically important crops such as Citrus, Malus, Rubus and Vitis (Hyde et al. 2013; Jayawardena et al. 2014; Fan et al. 2017). The species appear to have a narrow host range, usually limited to a single host (Fan et al. 2017). However, a few species have a broad host range e.g., E. anacardii, E. leucospermi, E. piri and E. viola.
Morphological based identification and diversity
Kirk et al. (2008) estimated that there are 48 species of Elsinoe and 52 species of Sphaceloma. There are 190 species epithets under Elsinoe and 169 epithets under Sphaceloma in Index Fungorum (Index Fungorum 2018). Most Elsinoe species described to date need to be recollected and epitypified. Fan et al. (2017) designated 13 epitypes based on taxonomy and phylogenetic data. In accordance with the “One Fungus, one name” concept, the sexual name Elsinoe was protected over Sphaceloma. Therefore, many names in Sphaceloma should be transferred to the genus Elsinoe. Fan et al. (2017) relocated 26 Sphaceloma species to Elsinoe. In their study, eight new species were introduced, leading to a total of 75 Elsinoe species supported by morphology and molecular data.
Colony and spore morphology are the primary characters to identify species of Elsinoe (Fan et al. 2017). Species have overlapping colony and spore characters making identification based on morphology difficult. Therefore, use of DNA sequence data is crucial in identifying these species.
Molecular based identification and diversity
The first molecular study on this genus was by Tan et al. (1996), who investigated the genetic differences among the citrus scab pathogens E. fawcettii and E. australis from South America and S. fawcettii var. scabiosa from Australia. The asexual morph and sexual morph relationship was resolved by Cheewangkoon et al. (2009) by analysing rDNA sequence data. Few molecular studies have been carried out on this genus. Schoch et al. (2006) and Boehm (2009) using rDNA data showed that the species of Elsinoe constitute a subclade among the species of Myriangiaceae. However, Schoch et al. (2006) used only four Elsinoe strains and one Myrangium strain. Swart et al. (2001), based on ITS sequence data, delineated six Elsinoe species associated with the scab disease of Proteaceae and proposed three new species. Similar studies (e.g., Summerbell et al. 2006; Everett et al. 2011) described species of this genus associated with other host plants. Everett et al. (2011) and Hyde et al. (2013) carried out higher level phylogenetic studies on Dothideomycetes, which included strains of Elsinoe. Jayawardena et al. (2014) using the available sequence data on ITS, LSU, SSU, RPB2 and TEF1-α in GenBank provided evidence that Elsinoaceae can be considered as a separate family within the order Myriangiales. At the time, 12 Elsinoe species were included in this analysis, but ex-type sequence data was available for only a few species. Most species are based on old specimens without sequence data (Jayawardena et al. 2014). Fan et al. (2017) used 119 isolates representing 67 host genera from 17 countries and analysed a combined multigene analysis (ITS, LSU RPB2 and TEF1-α) with 64 ex-type strains. However, Jayawardena et al. (2014) and Fan et al. (2017) were unable to include the generic type E. canavaliae due to a lack of DNA data. Even though there are several excellent studies on this genus associated with plant diseases, very few species have any available cultures or DNA data (Jenkins 1932 a, b, Bitancourt and Jenkins 1936). Therefore, epitypification from fresh collections is required to provide a stable and a workable taxonomy for this genus. This study reconstructs the phylogeny of Elsinoe based on a combined ITS, LSU, RPB2 and TEF1-α sequence data, updated with recently introduced species and it corresponds with previous studies.
Recommended genetic markers (Genus level) – ITS
Recommended genetic markers (Species level) – RPB2, TEF-1
Accepted number of species: There are more than 200 species epithets in Index Fungorum (2018) under this genus. However, only 75 species have molecular data are treated as accepted.
References: Hyde et al. 2013 (morphology, taxonomy), Jayawardena et al. 2014, Fan et al. 2017 (morphology, phylogeny), Chung and Liao 2008, Surup et al. 2018 (Phytotoxin).
Table Elsinoe. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate | ITS | LSU | RPB2 | TEF1-α |
| Elsinoe abutilonis | CBS 510.50* | KX887185 | KX886949 | KX887068 | KX886831 |
| E. ampelina | CBS 208.25 | KX887186 | KX886950 | KX887069 | KX886832 |
| E. anacardii | CBS 470.62* | KX887189 | KX886953 | KX887072 | KX886835 |
| E. annonae | CBS 228.64 | KX887190 | KX886954 | KX887073 | KX886836 |
| E. arachidis | CBS 511.50* | KX887191 | KX886955 | KX887074 | KX886837 |
| E. arrudai | CBS 220.50* | KX887194 | KX886958 | KX887077 | KX886840 |
| E. asclepiadea | CPC 18544* = RWB1202 = CBS 141937 | KX887195 | KX886959 | KX887078 | KX886841 |
| E. australis | CBS 314.32* | KX887198 | KX886962 | KX887081 | KX886844 |
| E. banksiicola | CBS 113734* = CPC1508 = CPC 1510 | KX887199 | KX886963 | KX887082 | KX886845 |
| E. barleriicola | CBS 471.62* = ATCC 14658 | KX887200 | KX886964 | KX887083 | KX886846 |
| E. bidentis | CBS 512.50* | KX887201 | KX886965 | KX887084 | KX886847 |
| E. brasiliensis | CPC 18528 = RWB 1133 | KX887204 | N/A | KX887087 | KX886850 |
| E. caleae | CBS 221.50* | KX887205 | KX886968 | KX887088 | KX886851 |
| E. centrolobii | CBS 222.50* | KX887206 | KX886969 | KX887089 | KX886852 |
| E. citricola | CPC 18535* = RWB 1175 | KX887207 | KX886970 | KX887090 | KX886853 |
| E. coryli | CBS 275.76* | KX887209 | KX886972 | KX887092 | KX886855 |
| E. diospyri | CBS 223.50* | KX887210 | KX886973 | KX887093 | KX886856 |
| E. embeliae | CBS 472.62* | KX887211 | KX886974 | N/A | KX886857 |
| E. erythrinae | CPC 18542* = RWB 1196 | KX887214 | KX886977 | KX887096 | KX886860 |
| E. eucalypticola | CBS 124765* = CPC 13318 | KX887215 | KX886978 | KX887097 | KX886861 |
| E. eucalyptorum | CBS 120084* = CPC 13052 | KX887216 | KX886979 | KX887098 | KX886862 |
| E. euphorbiae | CBS 401.63* | KX887217 | KX886980 | KX887099 | KX886863 |
| E. fagarae | CBS 514.50* | KX887218 | KX886981 | KX887100 | KX886864 |
| E. fawcettii | CBS 139.25* | KX887219 | KX886982 | KX887101 | KX886865 |
| E. fici | CBS 515.50 | KX887223 | KX886986 | KX887105 | KX886869 |
| E. fici-caricae | CBS 473.62* = ATCC 14652 | KX887224 | KX886987 | KX887106 | KX886870 |
| E. flacourtiae | CBS 474.62* = ATCC 14654 | KX887225 | KX886988 | KX887107 | KX886871 |
| E. freyliniae | CBS 128204* = CPC 18335 | KX887226 | KX886989 | KX887108 | KX886872 |
| E. genipae | CBS 342.39* | KX887227 | KX886990 | KX887109 | KX886873 |
| E. genipae-americanae | CBS 516.50* | KX887228 | KX886991 | KX887110 | KX886874 |
| E. glycines | CBS 389.64* | KX887229 | KX886992 | KX887111 | KX886875 |
| E. hederae | CBS 517.50* | KX887231 | KX886994 | KX887113 | KX886877 |
| E. ichnocarpi | CBS 475.62* = ATCC 14655 | KX887232 | KX886995 | KX887114 | KX886878 |
| E. jasminae | CBS 224.50* | KX887233 | KX886996 | KX887115 | KX886879 |
| E. jasminicola | CBS 212.63* | KX887234 | KX886997 | N/A | KX886880 |
| E. krugii | CPC 18531* = RWB 1151 | KX887235 | KX886998 | KX887116 | KX886881 |
| E. lagoa-santensis | CBS 518.50* | KX887239 | KX887002 | KX887120 | KX886885 |
| E. ledi | CBS 167.33* | KX887240 | KX887003 | KX887121 | KX886886 |
| E. lepagei | CBS 225.50* | KX887241 | KX887004 | KX887122 | A |
| E. leucospermi | CBS 111207* = CPC 1380 | KX887242 | KX887005 | KX887123 | KX886887 |
| E. lippiae | CBS 166.40* | KX887248 | KX887011 | KX887129 | KX886893 |
| E. mangiferae | CBS 226.50* | KX887249 | KX887012 | KX887130 | KX886894 |
| E. mattiroloanum | CBS 287.64 | KX887250 | KX887013 | KX887131 | KX886895 |
| E. menthae | CBS 322.37* | KX887253 | KX887016 | KX887134 | KX886898 |
| E. mimosa | CPC 19478* | KX887255 | KX887018 | KX887136 | KX886900 |
| E. oleae | CBS 227.59* | KX887256 | KX887019 | KX887137 | KX886901 |
| E. othonnae | CBS 139910* = CPC 24853 | N/A | N/A | N/A | N/A |
| E. perseae | CBS 406.34* | KX887258 | KX887021 | KX887139 | KX886903 |
| E. phaseoli | CBS 165.31* | KX887263 | KX887026 | KX887144 | KX886908 |
| E. piri | CBS 163.29 | KX887267 | KX887030 | KX887148 | KX886912 |
| E. pitangae | CBS 227.50* | KX887269 | KX887032 | KX887150 | KX886914 |
| E. poinsettiae | CBS 109333 | KX887270 | KX887033 | KX887151 | KX886915 |
| E. pongamiae | CBS 402.63* | KX887272 | KX887035 | KX887153 | KX886917 |
| E. populi | CBS 289.64 | KX887273 | KX887036 | KX887154 | KX886918 |
| E. proteae | CPC 1349* | N/A | N/A | N/A | N/A |
| E. protearum | CBS 113618* | KX887275 | KX887038 | KX887156 | KX886920 |
| E. punicae | CPC 19968 | KX887276 | KX887039 | KX887157 | KX886921 |
| E. quercus-ilicis | CBS 232.61* | KX887277 | KX887040 | N/A | KX886922 |
| E. randii | CBS 170.38* | KX887278 | KX887041 | KX887158 | KX886923 |
| E. rhois | CBS 519.50* | KX887280 | KX887043 | KX887160 | KX886925 |
| E. ricini | CBS 403.63 = ATCC 15030 | KX887281 | KX887044 | KX887161 | KX886926 |
| E. rosarum | CBS 212.33* | KX887283 | KX887046 | KX887163 | KX886928 |
| E. salicina | CPC 17824* | KX887286 | KX887049 | KX887166 | KX886931 |
| E. semecarpi | CBS 477.62* = ATCC 14657 | KX887287 | KX887050 | KX887167 | KX886932 |
| E. sesseae | CPC 18549 = RWB 1219 | KX887288 | KX887051 | KX887168 | KX886933 |
| E. sicula | CBS 398.59* | KX887289 | KX887052 | KX887169 | KX886934 |
| E. solidaginis | CBS 191.37* | KX887290 | KX887053 | KX887170 | KX886935 |
| Elsinoë sp. | CBS 128.14 | KX887291 | KX887054 | KX887171 | KX886936 |
| E. tectificae | CBS 124777* = CPC 14594 | KX887292 | KX887055 | KX887172 | KX886937 |
| E. terminaliae | CBS 343.39* | KX887293 | KX887056 | KX887173 | N/A |
| E. theae | CBS 228.50* | KX887295 | KX887058 | KX887175 | KX886939 |
| E. tiliae | CBS 350.73 = ATCC 24510 | KX887296 | KX887059 | KX887176 | KX886940 |
| E. veneta | CBS 164.29* = ATCC 1833 | KX887297 | KX887060 | KX887177 | KX886941 |
| E. verbenae | CPC 18561* = RWB 1232 | KX887298 | KX887061 | KX887178 | KX886942 |
| E. violae | CBS 336.35* | KX887302 | KX887065 | KX887182 | KX886946 |
| E. zizyphi | CBS 378.62* = ATCC 14656 | KX887303 | KX887066 | KX887183 | KX886947 |
| Myriangium hispanicum | CBS 247.33 | KX887304 | KX887067 | KX887184 | KX886948 |
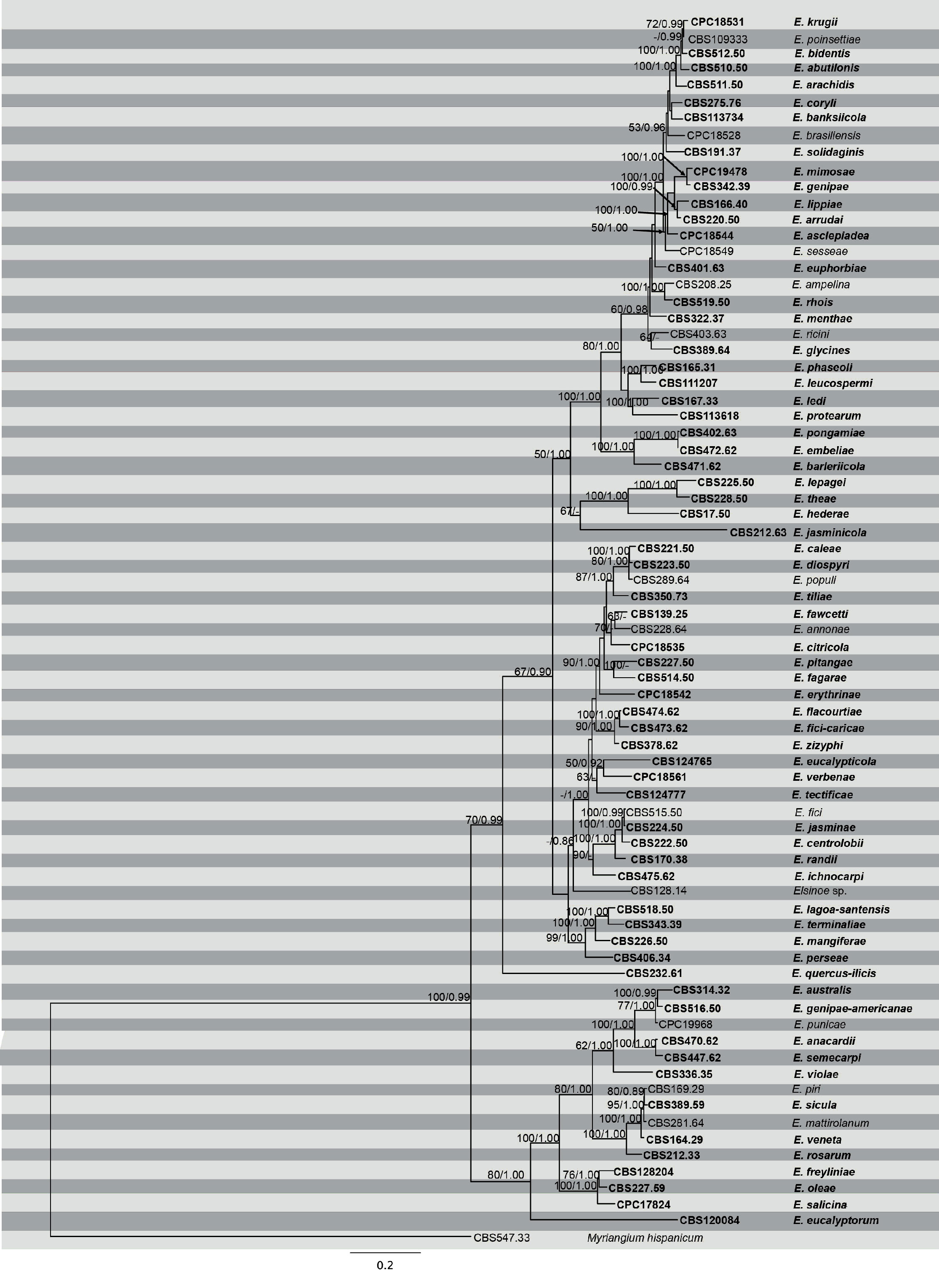
Phylogenetic tree generated by maximum Parsimony analysis of combined ITS, LSU, RPB2 and TEF1-α sequence data of Elsinoe species. Related sequences were obtained from GenBank. Seventy five strains are included in the analyses, which comprise 2479 characters including gaps. Single gene analyses were carried out (not shown) and the phylogeny generated were the same as combined analyses. Tree was rooted with Myriangium hispanicum (CBS 247.33). The maximum parsimonious dataset consisted of 1623 constant, 653 parsimony-informative and 203 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of ten equally most parsimonious trees with a length of 4748 steps (CI = 0.298, RI 0.699, RC = 0.208, HI = 0.702) in the first tree. Bayesian posterior probabilities and MP bootstrap values ≥ 50 % are shown respectively near the nodes. The scale bar indicates 0.2 changes. The ex-type strains are in bold.

No Comments