24 Oct Barriopsis
Barriopsis A.J.L. Phillips, A. Alves & Crous, in Phillips et al., Persoonia 21: 39 (2008).
Background
Stevens (1926) originally described the type species of Barriopsis in Physlospora as Physlospora fusca and Petrak and Deighton (1952) transferred it to Phaeobotryosphaeria. The fungus that was considered by Stevens (1926), and Petrak and Deighton (1952) did not have apiculi on its ascospores and was not similar to Phaeobotryosphaeria which had small, hyaline apiculi on the ascospores. von Arx and Müller (1954) considered Phaeobotryosphaeria as a synonym of Botryosphaeria. Based on morphological difference and molecular sequence data, Phillips et al. (2008) introduced Barriopsis. Species of Barriopsis are mostly saprobic and weak pathogens (Phillips et al. 2013).
Classification – Ascomycota, Dothideomycetes, Incertae sedis, Botryosphaeriales, Botryosphaeriaceae
Type species – Barriopsis stevensiana A.J.L. Phillips & Pennycook
Distribution – Species appear to be confined to regions with tropical or subtropical climates including Australia, Cuba, Iran and Thailand (Phillips et al. 2008; Abdollahzadeh et al. 2009; Liu et al. 2012; Phillips et al. 2013; Doilom et al. 2014; Konta et al. 2016; Dissanayake et al. 2016; Hyde et al. 2018b; Burgess et al. 2019).
Disease symptoms – Barriopsis species can be weak pathogens and their pathogenicities are uncertain (Phillips et al. 2008; Dissanayake et al. 2016). Barriopsis stevensiana and B. iraniana were isolated from infected branches, fruits and leaves with various disease symptoms, including dieback, canker, rot and necrosis, from Cupressus sempervirens, Mangifera indica, Citrus sp. and Olea sp. in northern and southern provinces of Iran (Abdollahzadeh et al. 2009). Species of this genus may be future emerging pathogens.
Hosts – Archontophoenix alexandrae, Cassia sp., Citrus sp., Mangifera indica, Olea sp. Tectona grandis (Phillips et al. 2008, 2013; Abdollahzadeh et al. 2009; Liu et al. 2012; Doilom et al. 2014; Konta et al. 2016; Dissanayake et al. 2016; Hyde et al. 2018b, 2020b).
Pathogen biology, disease cycle and epidemiology
Barriopisis in this article is considered as an emerging pathogen. Further studies to identify the biology, disease cycle and epidemiology are needed.
Morphological based identification and diversity
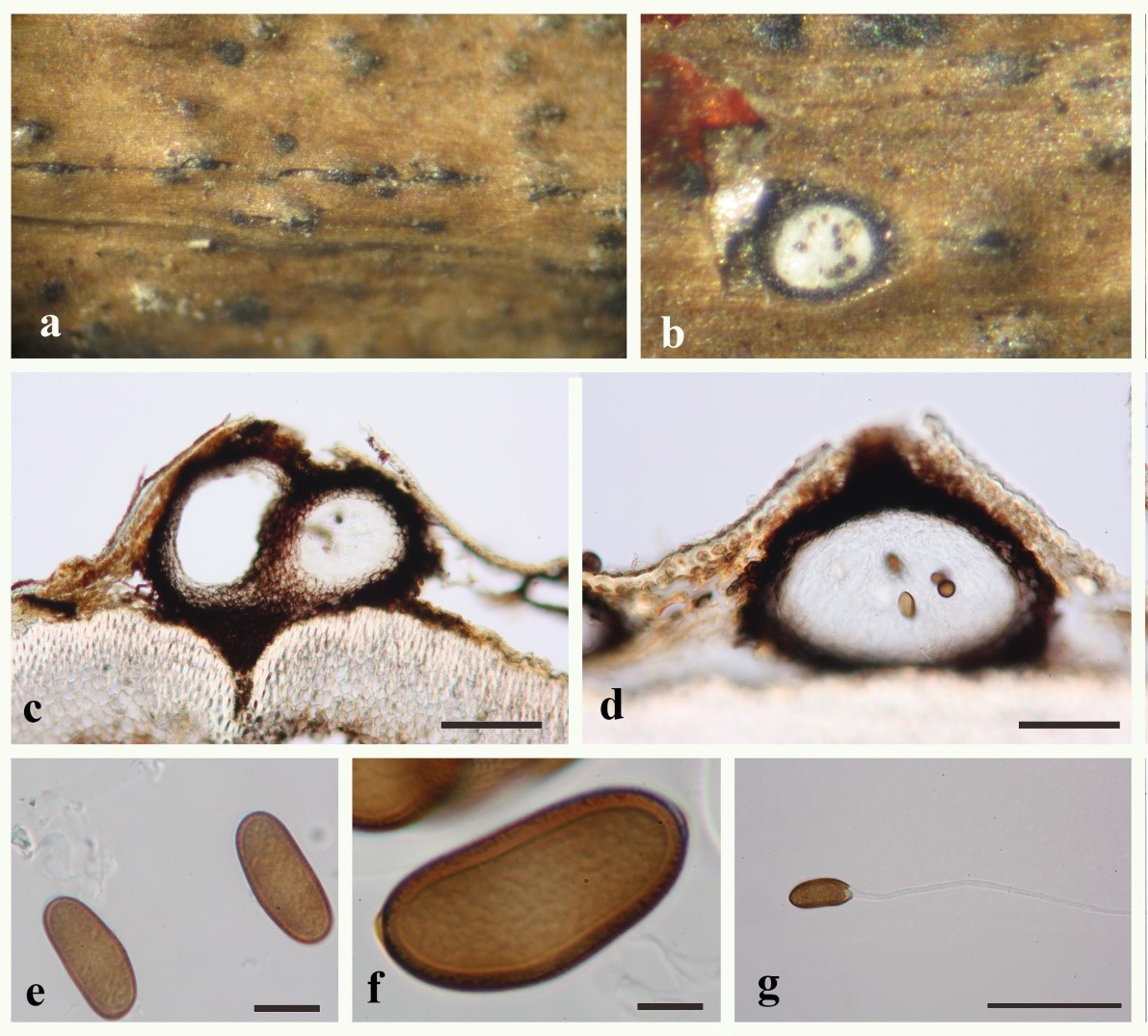
The sexual morph is characterized by brown aseptate ascospores that are widest in the center and lack terminal apiculi (Phillips et al. 2008, 2013; Doilom et al. 2014; Dissanayake et al. 2017; (Fig. 1)). Barriopsis archontophoenicis forms the sexual morph in culture medium after long periods of incubation (up to 6 months, Konta et al. 2016). The asexual morph is lasiodiplodia-like with hyaline conidia that become dark-brown and septate with irregular longitudinal striations (Stevens 1926). Abdollahzadeh et al. (2009) observed the asexual morphs of B. fusca and B. iraniana and confirmed that the morphology is similar to the description given by Stevens (1926). In their study, they revealed that this genus can be distinguished from other genera of Botryosphaeriaceae by the presence of visible striations on conidia at an early stage of development.
However, using morphology alone in identifying these species is not wise due to the overlapping of morphological characters within the genus. Therefore, the use of multi loci phylogeny along with morphology is recommended for this genus. Very little is known about the diversity and pathogenicity of this botryosphaeriaceous genus and future studies are needed to confirm its pathogenic nature.
Fig 1. Barriopsis stevensiana MFLU 19–1560. a Ascomata on dead twigs of Cassia sp. b Ascomata cut through horizontally showing the white contents with dark spots. c, d Sections through ascomata. e–f Ascospores. g Germinated ascospore. Scale bars: c, d = 200 µm, e–f = 20 µm, g = 100 µm.
Molecular based identification and diversity
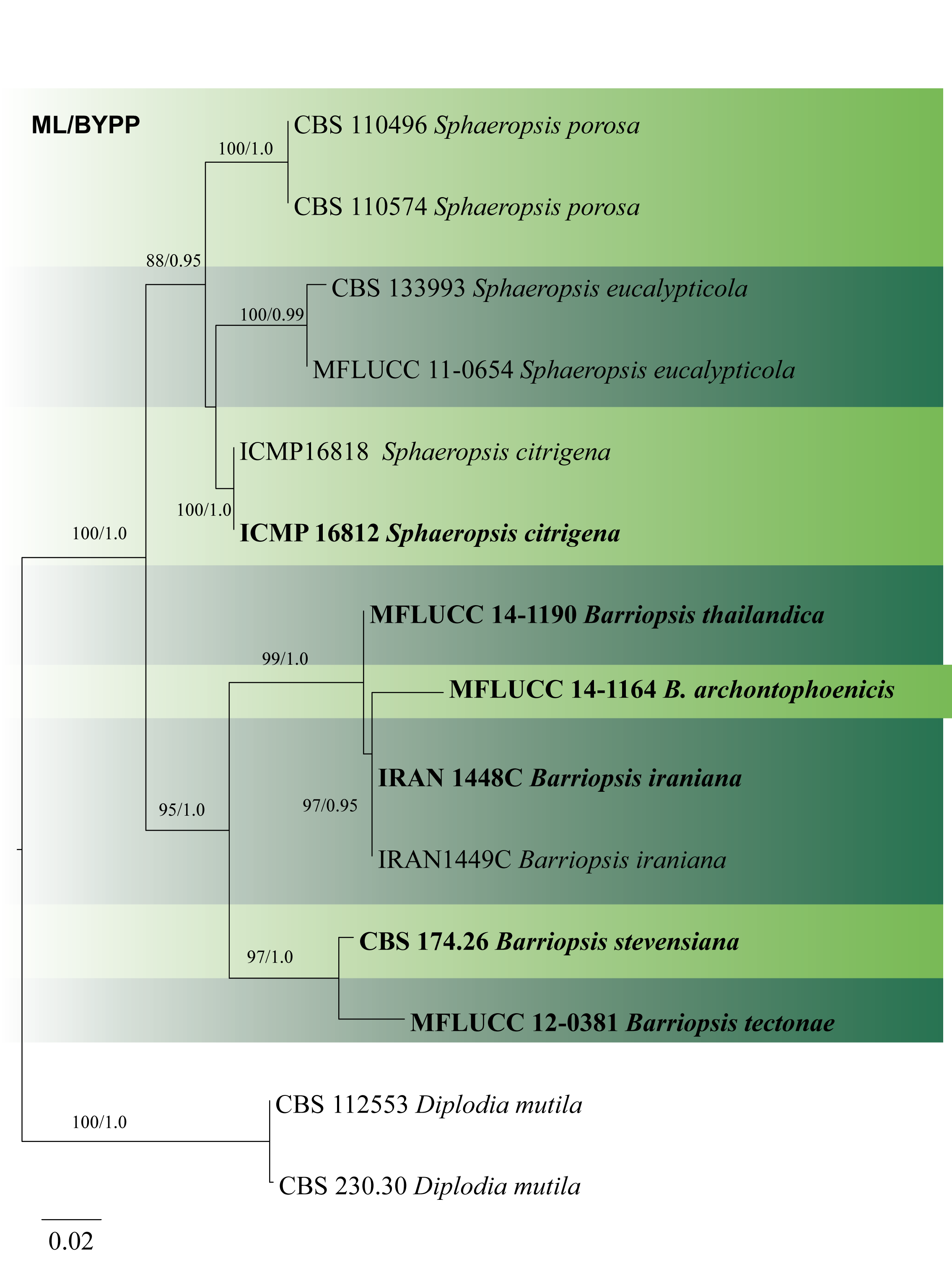
Phillips et al. (2008) using SSU, ITS, LSU, tef1 and tub2 sequence data established Barriopsis which is sister to Phaeobotryon. Based on ITS and tef1 sequence data, Abdollahzadeh et al. (2009) introduced B. iraniana. Doilom et al. (2014) introduced B. tectonae based on ITS, tub2 and tef1 sequence data. In this study, it was mentioned that ITS and tub2 sequence data have lesser variation, while tef1 sequence data have considerable variation. Konta et al. (2016) added a new species, B. archontophoenicis with the use of ITS, LSU, SSU and tef1 sequence data. In this study, we construct the phylogenetic tree for the accepted species based on ITS and tef1 sequence data (Fig. 2).
Recommended genetic marker (genus level) – ITS
Recommended genetic markers (species level) – tef1
Accepted number of species – There are six species epithets in Index Fungorum (2020), however only five species have DNA sequence data (Table 1).
References – Phillips et al. 2008, Abdollahzadeh et al. 2009 (morphology and phylogeny); Dissanayake et al. 2017 (accepted number of species, phylogeny); Doilom et al. 2014, Konta et al. 2016 (new species).
Table 1 DNA barcodes available for Barriopsis. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Isolate | ITS | tef1 |
| Barriopsis archontophoenicis | MFLUCC 14-1164* | KX235306 | KX235305 |
| B.iraniana | IRAN 1448C* | FJ919663 | FJ919652 |
| IRAN1449C | FJ919665 | FJ919654 | |
| B.tectonae | MFLUCC 12-0381* | KJ556515 | KJ556516 |
| B. thailandica | MFLUCC 14-1190* | KY115675 | KY115676 |
| B. stevensiana | CBS 174.26* | NR119698 | – |
Fig 2 Phylogram generated from maximum likelihood analysis based on combined ITS, and tef1 sequence data of Barriopsis species and closely related taxa. Fifteen strains are in the combined sequence analyses, which comprise 865 characters including gaps. Diplodia mutila (CBS 112553 andCBS 230.30) was used as the outgroup taxa. Tree topology of the ML analysis was similar to the one generated from BI. The best scoring RAxML tree with a final likelihood value of -2372.487246 is presented. The matrix had 201 distinct alignment patterns, with 12.30% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.207721, C = 0.288041, G = 0.271092, T = 0.233145; substitution rates AC = 1.068561, AG = 2.489613, AT = 0.682766, CG = 1.417925, CT = 4.236517, GT = 1.000000; gamma distribution shape parameter α= 1.343820.RAxML bootstrap support value ≥50% and BYPP ≥0.95 are shown respectively, near the nodes. Ex-type strains are in bold.

No Comments