20 Sep Botryosphaeria
Botryosphaeria Ces. & De Not., Comm. Soc. crittog. Ital. 1(fasc. 4): 211 (1863)
Background
The genus Botryosphaeria (Botryosphaeriaceae) was introduced by Cesati and de Notaris (1863), revised by Saccardo (1877) and is based on the type species Botryosphaeria dothidea (Barr 1972; Slippers et al. 2004c). This genus has undergone various revisions and updates over the years, at times encompassing a diverse range of morphologies. von Arx and Müller (1954) examined 183 taxa of Botryosphaeriales and reduced them to eleven species, with extensive synonymies under B. dothidea and B. quercuum, together with nine new combinations. In later studies, these synonymies were not always accepted (Shoemaker 1964; Sivanesan 1984; Slippers et al. 2004a; Phillips et al. 2009). Slippers et al. (2004b) epitypified the type species Botryosphaeria dothidea and provided an ex-epitype culture based on morphology and phylogeny of combined ITS, TEF1-α and TUB2 sequence data. This set a firm basis for the resolution of species.
Classification – Dothideomycetes, incertae sedis, Botryosphaeriales, Botryosphaeriaceae
Type species – Botryosphaeria dothidea (Moug.) Ces. & De Not., Comm. Soc. crittog. Ital. 1(fasc. 4): 212 (1863)
Distribution – Worldwide
Disease Symptoms – Leaf spots, cankers, dieback, fruit rot, gummosis, and even plant death.
Hosts – Plurivorous
Morphological based identification and diversity
Species in the genus Botryosphaeria have hyaline ascospores that can become pale brown with age, uniloculate ascomata often aggregated or forming botryose clusters, asexual morphs with thin-walled, aseptate, hyaline conidia that sometimes become olivaceous, occasionally forming one or two septa when aged, they typically lack a mucoid sheath and apical appendage. A search in MycoBank (July 2018) revealed 292 names in Botryosphaeria while Index Fungorum (July 2018) provided 279 names. However, only 13 species are known in culture. Dissanayake et al. (2016) included ten Botryosphaeria species in their phylogeny. Subsequently, Li et al. (2018) introduced three novel species, B. pseudoramosa, B. qingyuanensis and B. wangensis.
Colony and conidial morphology are the primary characters to identify species within this genus. Colonies are olivaceous becoming grey with reverse black. Mycelial mat is moderately dense. Conidia are narrowly fusiform, or irregularly fusiform, base sub-truncate to bluntly rounded (Phillips et al. 2013). However, we consider morphological characters alone are inadequate to identify species due to plasticity and overlapping of conidial dimensions.
Molecular based identification and diversity
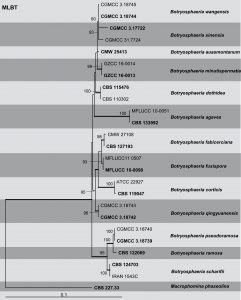
Recent advances in DNA-based molecular techniques have provided efficient tools to characterize and identify species in the genus Botryosphaeria (Slippers and Wingfield 2007; Phillips et al. 2013; Dissanayake et al. 2016; Li et al. 2018). Studies applying these tools are revealing significantly greater diversity on some hosts than was previously realized. Recent studies on the taxonomy of Botryosphaeria have employed molecular methods to reveal phylogenetic relationships among species and to resolve species complexes (Denman et al. 2003; Alves et al. 2004; Phillips et al. 2005). The present phylogenetic analysis was performed based on up to date ex-holotype or ex-epitype sequences available in GenBank. This study updates the phylogeny of Botryosphaeria based on combined analyses of ITS and TEF1- α sequence data (Table, Fig) and corresponds to the previous studies.
Recommended genetic markers (genus level) – SSU, LSU
Recommended genetic markers (species level) – ITS, TEF1 – α
Accepted number of species: There are 282 species epithets in Index Fungorum (2018) under this genus. However, only 13 are accepted.
References: Phillips et al. 2013 (morphology, phylogeny), Dissanayake et al. 2016 et al. (phylogeny).
Table Botryosphaeria. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate | ITS | TEF1-a | LSU |
| Botryosphaeria agaves | MFLUCC 11-0125* | JX646791 | JX646856 | JX646808 |
| B. agaves | MFLUCC 10-0051 | JX646790 | JX646855 | JX646807 |
| B. auasmontanum | CMW 25413* | KF766167 | EU101348 | KF766332 |
| B. corticis | CBS 119047* | DQ299245 | EU017539 | EU673244 |
| B. corticis | ATCC 22927 | DQ299247 | EU673291 | EU673245 |
| B. dothidea | CBS 115476* | AY236949 | AY236898 | DQ377852 |
| B. dothidea | CBS 110302 | AY259092 | AY573218 | EU673243 |
| B. fabicerciana | CBS 127193* | HQ332197 | HQ332213 | N/A |
| B. fabicerciana | CMW 27108 | HQ332200 | HQ332216 | N/A |
| B. fusispora | MFLUCC 10-0098* | JX646789 | JX646854 | JX646806 |
| B. fusispora | MFLUCC 11-0507 | JX646788 | JX646853 | JX646805 |
| B. minutispermatia | GZCC 16-0013* | KX447675 | KX447678 | N/A |
| B. minutispermatia | GZCC 16-0014 | KX447676 | KX447679 | N/A |
| B. qingyuanensis | CGMCC3.18742* | KX278000 | KX278105 | MF410042 |
| B. qingyuanensis | CGMCC3.18743 | KX278001 | KX278106 | MF410043 |
| B. ramose | CBS 122069* | EU144055 | EU144070 | N/A |
| B. scharifii | CBS 124703* | JQ772020 | JQ772057 | N/A |
| B. scharifii | CBS 124702 | JQ772019 | JQ772056 | N/A |
| B. pseudoramosa | CGMCC3.18739* | KX277989 | KX278094 | MF410031 |
| B. pseudoramosa | CGMCC3.18740 | KX277992 | KX278097 | MF410034 |
| B. sinensia | CGMCC 3.17722* | KT343254 | KU221233 | N/A |
| B. sinensia | CGMCC 3.17724 | KT343256 | KU221234 | N/A |
| B. wangensis | CGMCC3.18744* | KX278002 | KX278107 | MF410044 |
| B. wangensis | CGMCC3.18745 | KX278003 | KX278108 | MF410045 |
Fig: Phylogenetic tree generated by maximum likelihood analysis of combined ITS and TEF1-a sequence data of Botryosphaeria species. Related sequences were obtained from GenBank. Twenty-five strains are included in the analyses, which comprise 903 characters including gaps. The tree was rooted with Macrophomina phaseolina (CBS 227.33). Tree topology of the ML analysis was similar to the BI. The best scoring RAxML tree with a final likelihood value of – 2158.395527 is presented. The matrix had 528 distinct alignment patterns, with 17.64% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.210797, C = 0.294704, G = 0.256067, T = 0.238431; substitution rates AC = 0.448510, AG = 1.879010, AT = 0.864074, CG = 0.550809, CT = 3.948379, GT = 1.000000; gamma distribution shape parameter α = 0.387391. RAxML bootstrap support values ≥50% (BT) are shown respectively near the nodes. Bayesian posterior probabilities ≥0.95 (PP) indicated as thickened black branches. The scale bar indicates 0.1 changes. The ex-type strains are in bold.

No Comments