27 Oct Rhizopus
Rhizopus Ehrenb., Nova Acta Phys.-Med. Acad. Caes. Leop.-Carol. Nat. Cur. 10: 198 (1821)
Background
Rhizopus is classified in the subphylum Mucoromycotina, class Mucoromycetes, order Mucorales and family Rhizopodaceae (Wijayawardene et al, 2018, 2020). The genus is one of the most diverse and constitutes an important genus within the order Mucorales. Rhizopus species are common post-harvest pathogens of fruits, vegetables, crops and stored foods, while some Rhizopus species are human pathogens. Rhizopus arrhizus and Rhizopus microsporus can cause mucoromycosis in immunocompromised humans (Yildirim et al. 2010; Benedict and Brandt 2016). Morphology-based (size of sporangia and sporangiophores, and rhizoids) and physiology-based (growth temperature) identification and classification grouped the genus in three groups: R. microsporus, R. stolonifer, and R. arrhizus (syn: R. oryzae) (Schipper 1984). Schipper (1984) and Schipper and Stalpers (1984), provided the first significant monographs of Rhizopus. Fundamental morphological-based identification was provided which is still widely used in current taxonomic classification for Rhizopus (Schipper 1984; Schipper and Stalpers 1984; Hartanti et al. 2015). The inclusion of DNA-based phylogenetic tools has resulted in significant changes in the taxonomic classification (Vebliza et al. 2018). With the implementation of molecular-based identification, Abe et al. (2006, 2010), Zheng et al. (2007b), and Liu et al. (2007) provided significant contributions in the classification of Rhizopus. Briefly, in current taxonomy Rhizopus arrhizus is a synonym of R. oryzae, R. reflexus to R. lyococcus and Amylomyces rouxii is a synonym of Rhizopus arrhizus (Liu et al. 2007; Hyde et al. 2014; Vebliza et al. 2018).
Phylogenomic approaches have the potential to provide a clear understanding of the inter-relationships of species (Gryganskyi et al. 2018). In recent revisions, data from whole-genome sequencing have been used (Gryganskyi et al. 2018). Phylogenetic analysis based on a dataset of 192 orthologous protein-coding genes extracted from whole-genome sequencing of representative species provided a robust phylogeny and tree topology for Rhizopus. The phylogenetic analysis resulted in similar tree topology obtained from studies which utilize ITS and pyrG genes or 76 orthologous proteins from the genomes (Liu et al. 2007; Chibucos et al. 2016). In brief, R. microsporus is suggested to be a monophyletic sister clade to other Rhizopus clades, R. stolonifer was found to be sister to R. arrhizus and R. delemar and these four species are monophyletic (Gryganskyi et al. 2010, 2018)
A comparative analysis of the mating-type locus across Rhizopus revealed that its structure is flexible even between different species in the same genus, but shows similarities between Rhizopus and other mucoralean taxa. Variation of the genome size was also noted to be approximately three-fold within a species which are induced by changes in transposable element copy numbers and genome duplications (Gryganskyi et al. 2018). Bruni et al. (2019) successfully adapted the CRISPR-Cas 9 technique for inducing pyrF gene-specific mutations in two strains of R. delemar, the causative agent of mucoromycosis. This new tool is suggested to be useful in investigating the pathogenesis mechanisms of R. delemar and also generating specific mutants of Mucorales fungi.
Classification – Mucoromycota, Mucoromycotina, Mucoromycetes, Mucorales, Rhizopodaceae
Type species – Rhizopus stolonifera (Ehrenb.) Vuill. 1902
Distribution – worldwide
Disease symptoms – Rhizopus blight, Rhizopus head rot and Rhizopus soft rot
Rhizopus blight: Rhizopus blight can affect flowers, leaves, and stems. When infected, the plant shows symptoms such as soft and mushy brown rot. The rot produces white mycelia with black sporangia and the abundant mycelia projects a ‘bearded’ appearance. Spores of the fungus can be spread by water and air. The mode of infection is similar to bacterial soft rot in which enzymes secreted by the fungus causes cell deterioration of the host tissue. The fungi require high temperatures, high humidity and weakened host tissues or wounds (Hartley 1992).
Rhizopus head rot on sunflowers: Rhizopus head rot may be caused by several Rhizopus species such as Rhizopus arrhizus, R. microsporus and R. stolonifer (Markell et al. 2015). Historically, Rhizopus head rot was deemed as a minor disease. However, recent surveys have shown their severity. Initial signs of Rhizopus head rot are dark spots of different sizes on different types of wounds on the plant. Soft watery rot appears on the infected fruit which often turns dark brown and extends to the back of the flower head, sepals and peduncles as the disease progresses. The infected sunflower receptacle disintegrates and becomes soft and pulpy. Infection by Rhizopus causes the head to shrivel and dry. Morphological characteristics are mycelial strands bearing sporangiophore and sporangia which are seen as the disease advances (Markell et al. 2015; Zhou et al. 2018). These whiskers are tufts of hyphae containing numerous sporangia and generally appear around lenticels or breaks in the periderm. Sometimes hyphae may not be visible on the outside of the root but can be viewed by pulling apart the infected tissue, giving it a stringy appearance (Clark et al. 2013).
Rhizopus soft rot: Common causative agents of Rhizopus soft rot are Rhizopus stolonifer and Rhizopus oryzae. The disease is considered as one of the most common and destructive postharvest diseases in many plants such as sweet potato (Ipomoea batatas), potato (Solanum tuberosum) and tomato (Solanum lycopersicum). The most frequent mode of infection is wounds and injuries present on the plants. Studies have also shown that the type of wounding and storage time have a significant impact on the susceptibility of infection by Rhizopus species (Scruggs and Quesada-Ocampo 2016). Earliest symptoms of infections are soft water-soaked lesions. The disease spreads across the wounded area and progresses to the extremities of the substrate. Hyphae soon develop on the rotten tissues and produce grey sporangiophores which subsequently bear sporangia (Khokhar et al. 2019). Whiskers are characteristics features of Rhizopus soft rot and have been reported in the case of soft rot on sweet potatoes (Clark et al. 2013; Scruggs and Quesada-Ocampo 2016).
Hosts – Wide range of hosts including species of Allium, Ananas, Brassica, Cucumis, Cucurbita, Fragaria, Lycopersicon, Phaseolus, Pisum and Solanum (Farr and Rossman 2020)
Pathogen biology, disease cycle and epidemiology
The pathogen reproduces asexually. Spores of Rhizopus species are commonly found in the air and can survive easily on crop debris, fruits, vegetables, and even on tools and equipment. Factors such as the Rhizopus species, type of fruit, stage of maturity of the plant and fruit or the storage will have a slight difference in the disease cycle. Rhizopus stolonifer, as well as the other species causing post-harvest diseases such as Rhizopus soft rot, require wound injuries, cracks or any mechanical damage for entry (Hartley 1992; Bautista-Baños et al. 2014; Scruggs and Quesada-Ocampo 2016). Infection and colonization are highly dependent on the enzymes produced by the fungi. To establish within the host, Rhizopus species produce numerous enzymes, including amylase, pectinase, and cellulase that can damage cell walls and permit host colonization (Ogundero 1988; Tang et al. 2012). This results in the softening of the host tissue; one of the symptoms of the disease (Nelson 2009; Kwon et al, 2012; Bautista-Baños et al. 2014; Feliziani and Romanazzi 2016). During initial stages of infection, Rhizopus rot appears as water-soaked areas and in the case of Rhizopus stolonifer, the rot also exudes clear leachate. In the case of Rhizopus soft rot caused by R. oryzae in banana, the symptoms and disease cycle are similar to R. stolonifer (Kwon et al. 2012). In Okinawan sweet potatoes, the disease causes a soft and moist appearance and a stringy flesh during the initial stages and as the disease progresses, the tissue of the sweet potato turns brownish and eventually black (Nelson 2009). In the case of R. stolonifer, the fungal mycelia quickly spread across the infection site. The sporangia formed are normally black and the whole plant is covered by fungal mycelia (Bautista-Baños et al. 2014). The enzymes exuded from the pathogen generally liquefy the internal tissues, for an example in sweet potato parenchyma of the root becomes liquefied, leaving the periderm and outer fibres of the root intact (Scruggs and Quesada-Ocampo 2016). The disease becomes more severe in warm, humid environments (Zoffoli and Latorre 2011). Avoidance of Rhizopus species is difficult due to their ubiquitous nature; therefore, sanitation and storing produce under unfavourable disease conditions is the key to control this pathogen.
Morphology- based identification and diversity
Rhizopus is normally distinguished by rhizoids, stolons and single or branched sporangiophores (Vebliza et al. 2018). Identification of species takes into account the growth temperature, size of sporangiophore and sporangium and the branching of rhizoids (Abe et al. 2007). The white mycelia consist of coenocytic hyphae which bear the sporangiophore with normally black sporangia. These taxa are fast-growing and form rhizoids at the base of sporangiophores. The sporangium contains a columella and spores (Bullerman 2003). During the sexual stage, there is the formation of zygospores and chlamydospores can also be seen during the growth of the fungi (Bullerman 2003; Abe et al. 2007).
Molecular identification and diversity
Traditionally, Rhizopus species were classified using morphological characters such as the shape and size of the structures (chlamydospores, rhizoids, sporangiophores and columellae) and physiological features such as optimal growth conditions. Current classification and taxonomic grouping follow that of Schipper (Schipper 1984). Schipper classified Rhizopus into three groups namely R. microsporus, R. stolonifer and R. arrhizus based on the physiological factors and morphology (Abe et al. 2010; Grygansky et al. 2018). Later, studies such as Abe et al. (2006), Liu et al. (2007), Zheng et al. (2007), Abe et al. (2010) implemented molecular phylogeny using DNA sequence data in the classification of these fungi. With novel approaches used, the classification proposed by Schipper was found to agree with some recent studies while others divided the genus into ten species and seven varieties or eight species. Zheng et al. (2007b) used zygospore formation, and molecular systematic morphological characters, mating compatibility, physiology and molecular systematic to accept the division of the genus in ten species and seven varieties. Abe et al. (2010) also used the rDNA ITS gene region together with actin-1 and tef1, to reorganize the proposed taxonomy into eight species instead of ten species. One important data provided by this study was the problematic rDNA ITS region of R. americanus. It was discovered that R. sexualis var. americanus has three rDNA ITS gene regions which are distinct from each other. However, Liu et al. (2007) were not able to obtain all three rDNA ITS gene region instead they were able to amplify only one ITS region which was similar to that of Rhizopus oryzae. So, this led to the conclusion that R. americanus was phylogenetically different from R. sexualis.
Genetic markers (species and genus level) – ITS and rpb1
Genetic markers (higher-level phylogeny) – SSU, LSU and act
Accepted number of species – There are 152 species epithets in Index Fungorum (2020), however only 12 species have DNA sequence data (Table 1).
References – Bullerman 2003, Abe et al. 2007 (morphology); Abe et al. 2010, Grygansky et al. 2018, Vebliza et al. 2018 (morphology and phylogeny)
Table 1 DNA barcodes available for Rhizopus. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species name | Isolate no | SSU | ITS | LSU |
| Rhizopus americanus | CBS 340.62* | NG_062623 | HM999967 | NG_057873 |
| R. arrhizus# | CBS 111231 | JN206338 | ||
| CBS 544.80 | JN206337 | |||
| NRRL 1469 | DQ641279 | |||
| R. caespitosus | CBS 427.87* | NG_062622 | NR_137056 | NG_057871 |
| 33515 | AF115730 | DQ466604 | ||
| R. delemar | CBS 392.95 | MH862535 | MH874170 | |
| R. homothallicus | CBS336.62* | NG_062624 | HM999968 | NG_057870 |
| FSU2530 | KJ408537 | KJ408567 | KJ408554 | |
| R. koreanus | EML-HO95-1* | KU058194 | KU058202 | KU058196 |
| EML-HO95-2 | KU058195 | KU058203 | KU058197 | |
| R. lyococcus | strain FSU10053 | KJ408545 | KJ408562 | |
| CBS 319.35 | AB100449 | |||
| CBS 320.35 | JN206373 | JN206534 | ||
| R. microsporus# | CBS 699.68* | HM999970 | ||
| CBS 337.62 | AB250177 | JN206362 | MH869765 | |
| R. niveus# | IFO 4810 | DQ641284 | ||
| R. oryzae# | CBS 112.07* | NG_062621 | NR_103595 | NG_056282 |
| CBS 130146 | MH865585 | MH877020 | ||
| R. schipperae | CBS 138.95 | NR077174 | HM849672 | |
| ATCC 96514* | NG_064824 | DQ641323 | NG_059417 | |
| R. sexualis# | CBS 336.39* | NG_063011 | AB113017 | MH867536 |
| R. stolonifer# | CBS 389.95 | DQ641318 | ||
| SICAUCC 19-0001 | MN148534 | MN267051 | MN148530 |
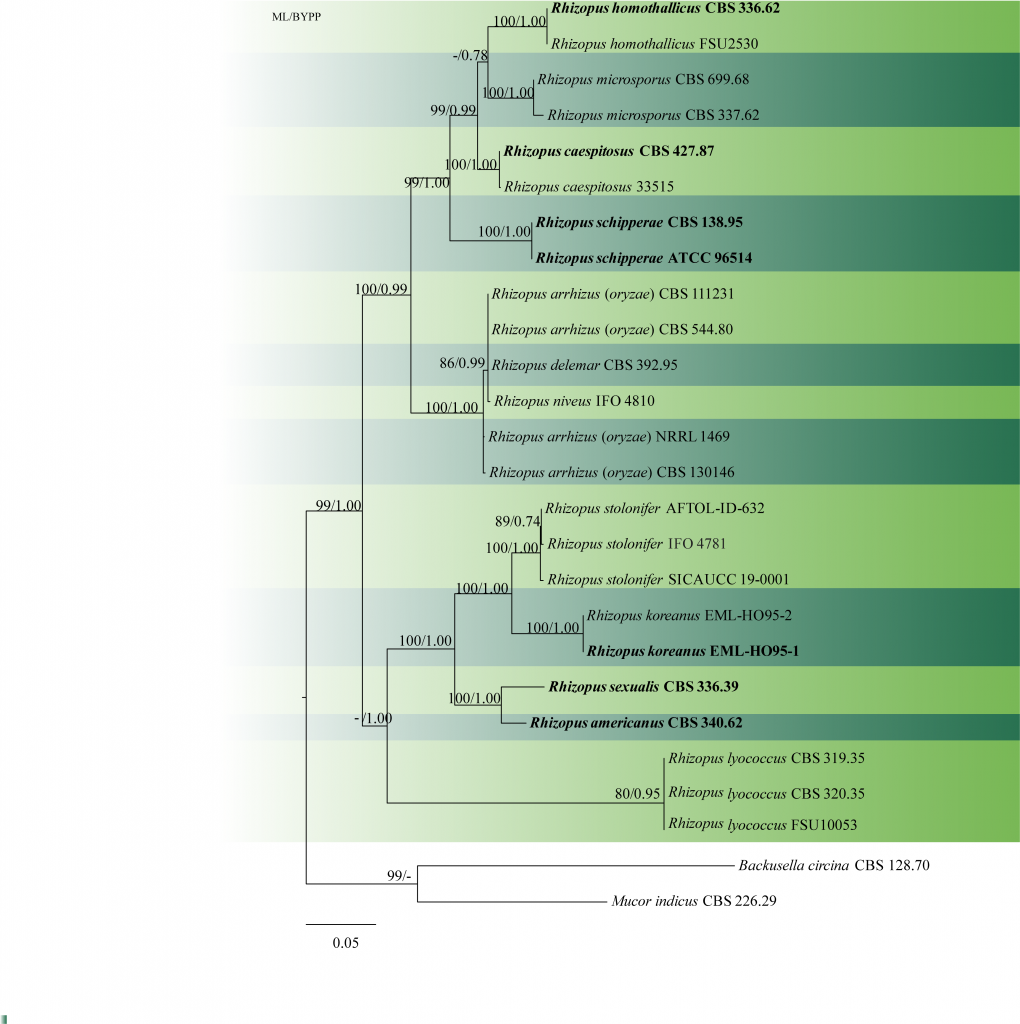
Fig. 1 Phylogenetic tree generated by maximum likelihood analysis of combined ITS-LSU-SSU sequence data of Rhizopus, Backusella and Mucor species. Twenty-six taxa containing 2600 characters including gaps were used in the phylogenetic analysis. The tree was rooted using Backusella circina (CBS 128.70) and Mucor indicus (CBS 226.29). The best scoring RAxML tree with a final likelihood value of -11138.113172is presented. The matrix contained 761distinct alignment patterns, with 41.20%of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.293528, C = 0.180262, G = 0.236482, T = 0.289728; substitution rates AC = 0.787730, AG = 2.127667, AT = 1.579251, CG = 0.715792, CT = 3.683423, GT = 1.000000; gamma distribution shape parameter α = 0.181944.ML bootstrap support values greater than 70% are shown near the nodes. The type species are in bold. Scale bar indicates the number of substitutions per site.

No Comments