15 Oct Fusarium
Fusarium Link, Mag. Gesell. naturf. Freunde, Berlin 3(1-2): 10 (1809)
Fusarium is a genus with 20 monophyletic species complexes (Rana et al. 2017). Species formerly belonged to F. solani species complex were transferred to genus Neocosmospora based on sexual morph characters and molecular phylogeny (Lombard et al. 2015; Sandoval-Denis & Crous (2018). Fusarium species are saprobes, parasites, endophytes, soil-borne or isolated from water (Rana et al. 2017). Species of Fusarium are economically important fungi as they are responsible for blights, cankers, rots, and wilts of horticultural, ornamental and forest crops in both agricultural and natural ecosystems, worldwide, and also human infections (Rana et al. 2017; Varela et al. 2013; Peraldi et al. 2013; Al-Hatmi et al. 2019; Maryani et al. 2019a, b). In nature, sexual morphs of Fusarium occur less commonly than the asexual morphs (Gräfenhan et al. 2011; Rossman et al. 1999).
Classification – Sordariomycetes, Hypocreomycetidae, Hypocreales, Nectriaceae
Type species – Fusarium sambucinum Fuckel, Hedwigia 2: 135.1863.
Distribution – Worldwide
Disease symptoms – blights, cankers, rots, and wilts
Plant pathogenic species of this genus have the capability to change their lifestyle to saprotrophic and can survive for long periods as chlamydospores in host tissues. Fusarium species damage their hosts by systemically colonizing and occluding the host xylem (Ploetz et al. 2003).
Hosts – Known from many host plant families.
Morphological based identification and diversity
Fusarium was also known from the sexual morphic fungus name Gibberella, which was suppressed in favour of Fusarium by Rossman et al. (2013). Currently, 1,552 Fusarium epithets are listed in Index Fungorum (2019). Variation and mutation in culture and lack of clear morphological characters for separating species are the main problems which make the species concept of Fusarium much broader (Geiser et al. 2004). It leads to the incorrect and confusing application of species names to toxigenic and pathogenic isolates (Geiser et al. 2004). Species boundaries have been inferred using multi-gene phylogenetic methods, reflecting the species diversity more than morphological treatments (Aoki and O’Donnell 1999; Geiser et al. 2004; O’Donnell 2000; O’Donnell et al. 1998a, b; Ward et al. 2002). A combined phylogenetic analysis of LSU, ITS, RPB2 and new phylogenetic marker acl1 by Gräfenhan et al. (2011), revealed that the early concept of Fusarium is not monophyletic. Fusarium sensu Wollenweber divided into two large groups, basal ‘Fusarium-like clades’, and the other one terminal ‘Fusarium clade’ in the Nectriaceae (Gräfenhan et al. 2011).
Fig. Sexual morph of a Fusarium sp. a Herbarium material. b Ascomata on the host. c Section of ascomata. d Section of the ostiolar region. e Peridium in face view. f–h Asci (h in Melzer’s reagent). i, j Ascospores. k Germinating ascospore. l, m Colony on MEA. Scale bars: c = 100 µm, d = 50 µm, e–k = 20 µm.
Molecular based identification and diversity
ITS and LSU are least informative in species-level identification of Fusarium (O’Donnell et al. 2008a; Hyde et al. 2014). Moreover, non-orthologous copies of the ITS2, which can lead to wrong phylogenetic inferences, can be detected in many species of Fusarium (Geiser et al. 2004; O’Donnell et al. 1998). Generally, for the species-level identification of fungi intron-rich regions of protein-coding genes are used as the markers (Geiser et al. 2004). The translation elongation factor 1-a (tef1), which lacks non-orthologous copies of the gene, is highly informative at the species level in Fusarium (Geiser et al. 2004). RPB1 and RPB2 are also very informative gene regions for species identification of Fusarium (O’Donnell et al. 2013; Hyde et al. 2014). Lombard et al. (2018) observed that tef1 and RPB2 genes provide better resolution of the species in the F. oxysporum complex than cmdA and tub2.
Recommended genetic markers (genus level) – ATP citrate lyase (acl1), tef1 and ITS
Recommended genetic markers (species level) – calmodulin-encoding gene (cmdA), tub2, tef1, RPB1 and RPB2
Accepted number of species: There are 1552 species epithets in Index Fungorum (2019) under this genus. More than 175 species have DNA sequence data.
References: Booth 1971, Rossman et al. 1999 (morphology), Rana et al. 2017, Gräfenhan et al. 2011; Laurence et al 2014; Lombard et al. 2018, 2019; Maryani et al. 2019a, b; Wang et al. 2019; Nalim et al. 2011(morphology, phylogeny).
Table Details of the Fusarium isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher No | RPB1 | RPB2 | tef1 |
| Fusarium anguioides | NRRL 25385*; ATCC 66485 | JX171511 | JX171624 | MH742689 |
| F. acaciae-mearnsii | NRRL 26755 | KM361640 | KM361658 | AF212449 |
| F. acuminatum | NRRL 36147; CBS 109232 | HM347174 | GQ505484 | GQ505420 |
| F. agapanthi | NRRL 54463* | KU900620 | KU900625 | KU900630 |
| F. albidum | NRRL 22152 | JX171492 | JX171605 | — |
| F. albosuccineum | NRRL 20459 | JX171471 | JX171585 | — |
| F. algeriense | NRRL 66647*; CBS 142638; IL-79 | MF120488 | MF120499 | — |
| F. ananatum | CBS 118516* | LT996188 | LT996137 | KU604416 |
| F. andiyazi | NRRL 31727; CBS 119857 | LT996189 | LT996138 | KP662901 |
| F. anthophilum | NRRL 13602; CBS 737.97 | LT996190 | LT996139 | KU711685 |
| F. arcuatisporum | CGMCC3.19493*; LC12147 | MK289799 | MK289739 | MK289584 |
| F. armeniacum | NRRL 6227 | JX171446 | JX171560 | HM744692 |
| F. asiaticum | NRRL 13818*; CBS 110257 | JX171459 | JX171573 | AF212451 |
| F. avenaceum | NRRL 54939 | JX171551 | JX171663 | MH582391 |
| F. aywerte | NRRL 25410 | JX171513 | JX171626 | KU171717 |
| F. babinda | NRRL 25539; CBS 396.96 | JX171519 | JX171632 | MH742712 |
| F. begoniae | CBS 403.97*; NRRL 25300 | LT996191 | LT996140 | AF160293 |
| F. beomiforme | NRRL 25174; CBS 740.97 | — | JX171619 | — |
| F. brachygibbosum | NRRL 13829 | JX171460 | JX171574 | — |
| F. buharicum | NRRL 13371; CBS 796.70 | JX171449 | JX171563 | — |
| F. bulbicola | NRRL 13618*; CBS 220.76 | KF466394 | KF466404 | KF466415 |
| F. burgessii | CBS 125537*; RBG 5319 | KJ716217 | HQ646393 | HQ667149 |
| F. callistephi | CBS 187.53* | — | MH484905 | MH484966 |
| F. carminascens | CBS 144738*; CPC 25800 | — | MH484937 | MH485028 |
| F. celtidicola | MFLUCC 16-0751* | — | MH576580 | — |
| F. citri | CGMCC3.19467*; LC6896 | MK289828 | MK289771 | MK289617 |
| F. citricola | CBS 142421*; CPC 27805 | LT746290 | LT746310 | LT746197 |
| F. coicis | NRRL 66233*; RBG5368 | KP083269 | KP083274 | KP083251 |
| F. commune | NRRL 28387 | — | JX171638 | KU171720 |
| F. concentricum | NRRL 25181*; CBS 450.97 | LT996192 | JF741086 | AF160282 |
| F. concolor | NRRL 13459*; CBS 961.87 | JX171455 | JX171569 | MH742681 |
| F. contaminatum | CBS 114899* | — | MH484901 | MH484992 |
| F. continuum | F201128 | KM520389 | KM236780 | KM236720 |
| F. convolutans | CBS 144207*; CPC 33733 | LT996193 | LT996141 | LT996094 |
| F. cugenangense | InaCCF984* | LS479560 | LS479308 | — |
| F. culmorum | NRRL 25475; CBS 417.86 | JX171515 | JX171628 | KY873384 |
| F. cuneirostrum | NRRL 31104 | — | EU329558 | EF408413 |
| F. curvatum | CBS 238.94; NRRL 26422; PD 94/184 | — | MH484893 | MH484984 |
| F. cyanostomum | NRRL 53998 | JX171546 | JX171658 | — |
| F. dactylidis | NRRL 29298* | KM361654 | KM361672 | DQ459748 |
| F. denticulatum | NRRL 25302; CBS 735.97 | LT996195 | LT996143 | AF160269 |
| F. desaboruense | InaCC F950 | LS479870 | LS479852 | — |
| F. dlaminii | NRRL 13164*; CBS 119860 | KU171681 | KU171701 | MK639039 |
| F. duoseptatum | InaCC F916* | LS479495 | LS479239 | — |
| F. elaeidis | CBS 217.49*; NRRL 36358 | — | MH484870 | MH484961 |
| F. enterolobii | CPC 27190 | — | LT746312 | LT746199 |
| F. equiseti | NRRL 13405 | — | GQ915491 | GQ915507 |
| F. fabacearum | CBS 144743*; CPC 25802 | — | MH484939 | MH485030 |
| F. ficicrescens | CBS 125178* | — | KT154002 | KP662899 |
| F. flocciferum | NRRL 25473 | JX171514 | JX171627 | — |
| F. foetens | NRRL 38302 | JX171540 | JX171652 | GU170559 |
| F. fracticaudum | CMW 252374 | LT996196 | LT996144 | KJ541059 |
| F. fractiflexum | NRRL 28852* | — | LT575064 | AF160288 |
| F. fredkrugeri | CBS 144209*; CPC 33747 | LT996199 | LT996147 | LT996097 |
| F. fujikuroi | NRRL 13566 | JX171456 | JX171570 | — |
| F. gaditjirrii | NRRL 45417 | — | JX171654 | KU171724 |
| F. gamsi | CBS 143610*; CPC 30862; OrSaAg4 | — | LT970760 | LT970788 |
| F. globosum | NRRL 26131*; CBS 428.97 | KF466396 | KF466406 | KF466417 |
| F. glycines | CBS 144746*; CPC 25808 | — | MH484942 | MH485033 |
| F. gossypinum | CBS 116613* | — | MH484909 | MH485000 |
| F. graminearum | NRRL 31084; CBS 123657 | JX171531 | JX171644 | HM744693 |
| F. grosmichelii | InaCC F833* | LS479548 | LS479295 | LS479744 |
| F. guilinense | CGMCC3.19495*; LC12160 | MK289831 | MK289747 | MK289594 |
| F. guttiforme | NRRL 22945 | JX171505 | JX171618 | — |
| F. hainanense | CGMCC3.19478*; LC11638 | MK289833 | MK289735 | MK289581 |
| F. heterosporum | NRRL 20693; CBS 720.79 | JX171480 | JX171594 | — |
| F. hexaseptatum | InaCC F866* | — | LS479359 | LS479805 |
| F. hoodiae | CBS 132474* | — | MH484929 | MH485020 |
| F. hostae | NRRL 29889 | JX171527 | JX171640 | AY329034 |
| F. inflexum | NRRL 20433 | — | JX171583 | AF008479 |
| F. ipomoeae | CGMCC3.19496*; LC12165 | MK289859 | MK289752 | MK289599 |
| F. iranicum | CBS 143608*; CPC 30860; OrSaAg2 | — | LT970757 | LT970785 |
| F. irregulare | CGMCC3.19489*; LC7188 | MK289863 | MK289783 | MK289629 |
| F. kalimantanense | InaCC F917* | LS479497 | LS479241 | LS479690 |
| F. konzum | CBS 119849* | LT996200 | LT996148 | LT996098 |
| F. kotabaruense | InaCC F963* | LS479875 | LS479859 | LS479445 |
| F. kuroshium | CBS 142642*; UCR3641 | KX262236 | KX262256 | KX262216 |
| F. kyushuense | NRRL 25349 | — | GQ915492 | GQ915508 |
| F. lacertarum | NRRL 20423; CBS 130185 | HM347137 | JX171581 | GQ505593 |
| F. lactis | NRRL 25200*; CBS 411.97 | LT996201 | LT996149 | AF160272 |
| F. langsethiae | NRRL 54940 | JX171550 | JX171662 | — |
| F. languescens | CBS 645.78*; NRRL 36531 | — | MH484880 | MH484971 |
| F. lateritium | NRRL 13622 | JX171457 | JX171571 | AY707173 |
| F. libertatis | CBS 144749*; CPC 28465 | — | MH484944 | MH485035 |
| F. longipes | NRRL 13368 | JX171448 | JX171562 | — |
| F. luffae | CGMCC3.19497*; LC12167 | MK289869 | MK289754 | MK289601 |
| F. lumajangense | InaCC F872* | — | LS479850 | LS479441 |
| F. lyarnte | NRRL 54252; CBS 125536 | JX171549 | JX171661 | — |
| F. mangiferae | NRRL 25226*; BBA 69662 | JX171509 | JX171622 | — |
| F. miscanthi | NRRL 26231 | — | JX171634 | KU171725 |
| F. mundagurra | NRR L66235*; RBG5717 | KP083272 | KP083276 | MK639058 |
| F. nanum | CGMCC3.19498*; LC12168 | MK289871 | MK289755 | MK289602 |
| F. napiforme | CBS 748.97*; NRRL 13604 | HM347136 | EF470117 | KU604409 |
| F. nelsonii | NRRL 13338 | JX171447 | JX171561 | GQ505402 |
| F. newnesense | NRRL 66237; RBG5443 | KP083271 | KP083277 | KJ397074 |
| F. nirenbergiae | CBS 840.88* | — | MH484887 | MH484978 |
| F. nisikadoi | NRRL 25179; CBS 742.97 | JX171507 | JX171620 | — |
| F. nurragi | NRRL 36452; CBS 392.96 | JX171538 | JX171650 | — |
| F. nygamai | NRRL 13448*; CBS 749.97 | LT996202 | EF470114 | AF160273 |
| F. odoratissimum | InaCC F822* | LS479618 | LS479386 | — |
| F. oligoseptatum | NRRL 62579*; FRC S-2581; MAFF 246283; CBS 143241 | KC691596 | KC691656 | KC691538 |
| F. oxysporum | CBS 144134* | — | MH484953 | MH485044 |
| F. palustre | NRRL 54056* | KT597718 | KT597731 | — |
| F. paranaense | CML1830* | — | KF680011 | KF597797 |
| F. parvisorum | CBS 137236*; FCC 5407; CMW 25267 | — | LT996150 | KJ541060 |
| F. pernambucanum | MUM 1862*; URM 7559 | MH668869 | — | — |
| F. petersiae | CBS 143231* | MG386139 | MG386150 | MG386159 |
| F. pharetrum | CBS 144751*; CPC 30824 | — | MH484952 | MH485042 |
| F. phialophorum | InaCC F971* | LS479545 | LS479292 | LS479741 |
| F. phyllophilum | NRRL 13617*; CBS 216.76 | KF466399 | KF466410 | KF466421 |
| F. pisi | NRRL 22278 | — | EU329501 | AF178337 |
| F. poae | NRRL 13714 | JX171458 | JX171572 | — |
| F. praegraminearum | NRRL 39664* | KX260125 | KX260126 | KX260120 |
| F. proliferatum | NRRL 22944; CBS 217.76 | JX171504 | JX171617 | — |
| F. pseudensiforme | NRRL 46517 | KC691615 | KC691645 | KC691555 |
| F. pseudocircinatum | NRRL 22946*; CBS 449.97 | LT996204 | LT996151 | AF160271 |
| F. pseudograminearum | NRRL 28062*; CBS 109956 | JX171524 | JX171637 | AF212468 |
| F. pseudonygamai | NRRL 13592*; CBS 417.97 | LT996205 | LT996152 | AF160263 |
| F. purpurascens | InaCC F886* | — | LS479385 | LS479827 |
| F. ramigenum | NRRL 25208*; CBS 418.98 | KF466401 | KF466412 | KF466423 |
| F. redolens | NRRL 22901; CBS 743.97 | — | KU171708 | KU171728 |
| F. riograndense | HCF3* | — | KX534003 | KX534002 |
| F. roseum | NRRL 22187 | JX171493 | JX171606 | — |
| F. sacchari | NRRL 13999; CBS 223.76 | JX171466 | JX171580 | KU711669 |
| F. salinense | CBS 142420*; CPC 26973 | LT746286 | LT746306 | LT746193 |
| F. sangayamense | InaCC F960* | LS479537 | LS479283 | — |
| F. sarcochroum | NRRL 20472; CBS 745.79 | JX171472 | JX171586 | — |
| F. scirpi | NRRL 13402 | JX171452 | JX171566 | GQ505592 |
| F. sibiricum | NRRL 53430* | — | HQ154472 | HM744684 |
| F. siculi | CBS 142422*; CPC 27188 | LT746299 | LT746327 | LT746214 |
| F. sororula | CBS 137242* | LT996206 | LT996153 | — |
| F. sporotrichioides | NRRL 3299 | JX171444 | JX171558 | HM744665 |
| F. staphyleae | NRRL 22316 | JX171496 | JX171609 | MH582426 |
| F. stercicola | CBS 142481*; DSM 106211 | — | KY556552 | KY556524 |
| F. stilboides | NRRL 20429; ATCC 15662 | JX171468 | JX171582 | — |
| F. subglutinans | NRRL 22016*; CBS 747.97 | JX171486 | JX171599 | HM057336 |
| F. sublunatum | NRRL 13384*; CBS 189.34 | JX171451 | JX171565 | — |
| F. subtropicale | NRRL 66764*; CBS 144706 | MH706972 | MH706973 | MH706974 |
| F. succisae | NRRL 13613; CBS 219.76 | LT996207 | LT996154 | AF160289 |
| F. sudanense | CBS 454.97*; NRRL 25451 | LT996208 | LT996155 | KU711697 |
| F. sulawesiense | InaCC F940* | — | LS479855 | LS479443 |
| F. tanahbumbuense | InaCC F965* | LS479877 | LS479863 | LS479448 |
| F. tardichlamydosporum | InaCC F958* | — | LS479280 | LS479729 |
| F. tardicrescens | NRRL 36113* | LS479474 | LS479217 | LS479665 |
| F. terricola | CBS 483.94* | LT996209 | LT996156 | KU711698 |
| F. thapsinum | NRRL 22045; CBS 733.97 | JX171487 | JX171600 | AF160270 |
| F. tjaetaba | NRRL66243*; RBG5361 | KP083267 | KP083275 | KP083263 |
| F. tjaynera | NRRL66246*; RBG5367 | KP083268 | KP083279 | — |
| F. torreyae | NRRL 54149 | JX171548 | HM068359 | HM068337 |
| F. torulosum | NRRL 22748; NRRL 13919 | JX171502 | JX171615 | — |
| F. transvaalense | CBS 144211*; CPC 30923 | LT996210 | LT996157 | LT996099 |
| F. tricinctum | NRRL 25481*; CBS 393.93 | JX171516 | HM068327 | MH582379 |
| F. triseptatum | CBS 258.50*; NRRL 36389 | — | MH484873 | MH484964 |
| F. udum | NRRL 22949; CBS 178.32 | LT996220 | LT996172 | AF160275 |
| F. venenatum | CBS 458.93* | — | KM232382 | KM231942 |
| F. verrucosum | NRRL 22566, BBA 64786 | — | JX171613 | — |
| F. verticillioides | NRRL 20956 | JX171485 | JX171598 | — |
| F. veterinarium | CBS 109898*; NRRL 36153 | — | MH484899 | MH484990 |
| F. volatile | CBS 143874* | — | LR596006 | LR596007 |
| F. witzenhausenense | CBS 142480* | — | KY556553 | KY556525 |
| F. xylarioides | NRRL 25486; CBS 258.52 | JX171517 | JX171630 | AY707136 |
| F. caatingaense | MUM 1859*; URM 6779 | MH668845 | LS398495 | LS398466 |
| F. goolgardi | NRRL 66250*; RBG5411 | KP083270 | KP083280 | KP101123 |
| F. humuli | CGMCC3.19374*; CQ1039 | MK289840 | MK289724 | MK289570 |
| Fusicolla aquaeductuum | NRRL 20686* | JX171476 | JX171590 | — |
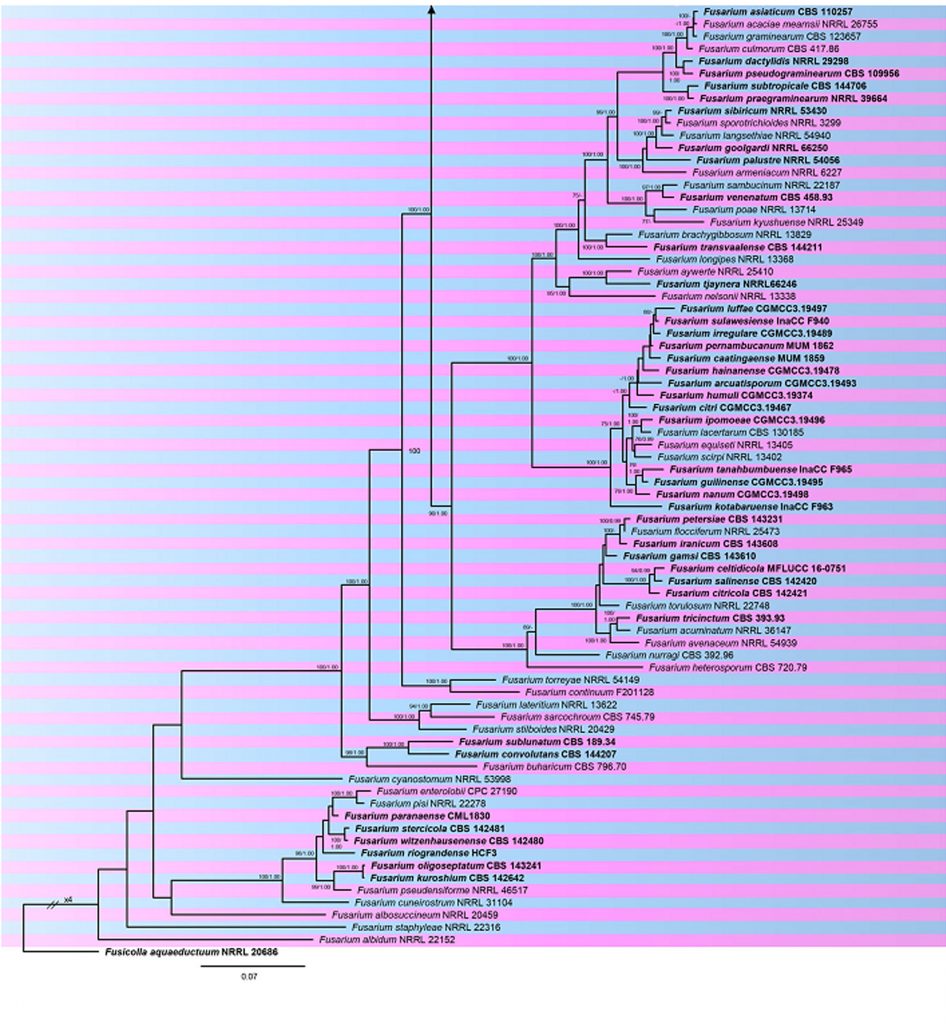
Fig. Phylogram generated from RAxML analysis based on combined RPB1, RPB2 and tef1 sequences of accepted species of Fusarium. Related sequences were obtained from GenBank. One hundred sixty-five taxa are included in the analyses, which comprise 3980 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted in Fusicolla aquaeductuum (NRRL 20696). Tree topology of the ML analysis was similar to the BYPP. The best scoring RAxML tree with a final likelihood value of -63956.723151 is presented. The matrix had 2254 distinct alignment patterns, with 25.13% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.257298, C = 0.251723, G = 0.248896, T = 0.242083; substitution rates AC = 1.366599, AG = 4.760724, AT = 1.266971, CG = 0.903914, CT = 9.564945, GT = 1.000000; gamma distribution shape parameter α = 1.089091. RAxML and maximum parsimony bootstrap support value ≥70% are shown near the nodes. The scale bar indicates 0.07 changes. The ex-type strains are in bold.



Josh Anderson
Posted at 15:14h, 10 OctoberI’d like to see if there could be a disucssion on the perspectives of this article regarding Fusarium solani.
“No to Neocosmospora: phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus” Fusarium. mSphere 5:e00810-20. https://doi.org/10.1128/mSphere.00810-20.