20 Sep Curvularia
Curvularia Boedijn, Bull. Jard. Bot. Buitenzorg, 3 Sér. 13: 123 (1933)
For synonyms see Index Fungorum (2018)
Background
The cosmopolitan Curvularia consists of pathogens and saprobes of various plants, as well as opportunistic pathogens of humans and animals. They are abundantly found as the pathogens of family Poaceae (Hyde et al. 2014). Curvularia lunata, C. trifolii and C. tuberculata can cause leaf spots and leaf blights of some cereal crops such as maize, rice and horticultural crops (Bermuda grasses and turf grasses) (de Luna et al. 2002; Hyde et al. 2014). The most frequent human and animal pathogens within the genus are C. aeria, C. borreriae, C. geniculata, C. inaequalis, C. lunata and C. verrucosa (Hyde et al. 2014). Curvularia is morphologically characterized by its dark mycelium, geniculate conidiophores with sympodial, tretic conidiogenous cells, conidia with smooth to slightly verrucose wall and several false septa (distosepta) (Hyde et al. 2014). The taxonomy of Curvularia has been well studied in recent years, however, the broadly perceived classification was redefined by Manamgoda et al. (2012, 2014) based on the phylogenetic relationships of ex-type strains of Curvularia species and recently collected Curvularia cultures from northern Thailand. See Hyde et al. (2014) and Marin-Felix et al. (2017) for further details.
Classification – Dothideomycetes, Pleosporomycetidae, Pleosporales, Pleosporaceae
Type species – Curvularia lunata (Wakker) Boedijn, Bull. Jard. bot. Buitenz, 3 Sér. 13(1): 127 (1933)
Distribution – Worldwide
Disease Symptoms – Leaf spots and blights, melting out, foot and root rot
The first symptoms to appear on leaves are elliptical brown spots. Gradually the spots enlarge and change colour to brownish black. In melting out disease symptoms on grasses may vary depending on the extent and severity of the attack. The first symptoms are seen as small spots with purple and black specks. Melting out starts as a leaf spot which will expands to the plant base and attacks the roots and crown. Leaf wilts, necrotic roots and plant death can be seen in a foot or root rot (Verma and Gupta 2009; Sunpapao et al. 2014).
Hosts – Mainly found on members of Poaceae. Also occurs on Actinidaceae, Aizoaceae, Caricaceae, Convolvulaceae, Fabaceae, Iridaceae, Lamiaceae, Lythraceae, Oleaceae, Polygonaceae, Rubiaceae and Vitaceae (Farr and Rossman 2018).
Morphological based identification and diversity
Species of Curvularia are traditionally characterized by dark mycelium, geniculate conidiophores with sympodial, tretic conidiogenous cells and elongated conidia. The conidia are smooth to tuberculate-walled, with several false septa (distosepta) and straight or curved due to an enlarged middle cell that is often more pigmented than the other cells (da Cunha et al. 2013). However, taxonomic classification of Curvularia spp. based exclusively on morphological characteristics was insufficient for designating new species because of their phenotypic variability and this has resulted in an inadequate understanding of curvularia-like species. Currently, there are 156 species epithets in Index Fungorum (www.indexfungorum.org; retrieved 24 March 2018) but most of these past records lack molecular data and comprehensive morphological descriptions. In a recent study, Marin-Felix et al. (2017) included 74 accepted Curvularia species to their phylogenetic analyses. Later Hyde et al. (2017) introduced Curvularia palmicola as another species and therefore, we constructed a tree with 75 Curvularia species.
Species delimitation in Curvularia based on morphology only is difficult with overlapping morphological characters among many species (Manamgoda et al. 2014, 2015), as also observed in Bipolaris (see under Bipolaris).
Molecular based identification and diversity
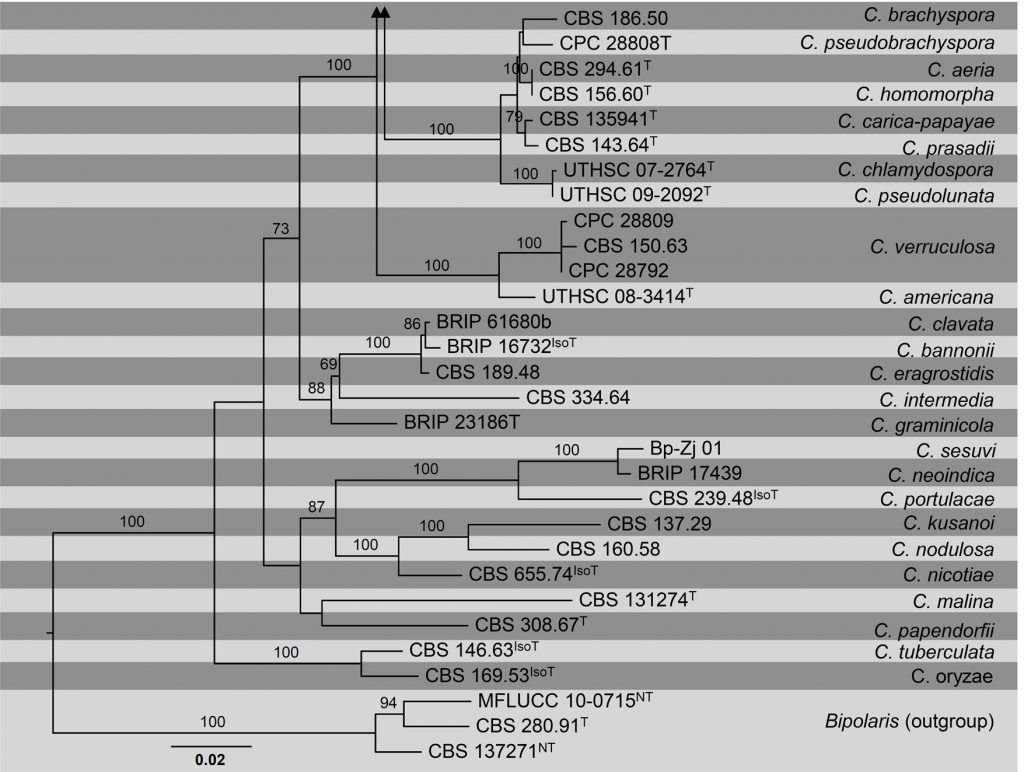
To achieve proper generic and species delimitation, phylogenetic studies using ITS, GAPDH and TEF1-α sequence data were recently performed (Manamgoda et al. 2012, 2014, 2015; Hyde et al. 2014, 2017; Marin-Felix et al. 2017). Phylogenetic studies based on these loci made it possible to reallocate species of Cochliobolus (sexual morph) to either Bipolaris or Curvularia (Marin-Felix et al. 2017). We update the phylogeny of this genus based on a combined ITS GAPDH and TEF1-α sequence data obtained from available ex-epitype, ex-isotype, ex-isolectotype, ex-paratype, ex-syntype and ex-type strains cultures (Table, Fig). The topological structure is in accordance with previous studies.
Recommended genetic markers (genus level) – LSU
Recommended genetic markers (species level) – GDPH
It is recommended to use a combination of ITS GAPDH and TEF (Manamgoda et al. 2015).
Accepted number of species: There are 399 species epithets in Index Fungorum (2018) under this genus. However, only 80 are accepted.
References: Sivanesan 1977 (morphology and pathogenicity), Manamgoda et al. 2011 (pathogenicity), Hyde et al. 2014, Marin-Felix et al. 2017 (morphology and phylogeny), Manamgoda et al. 2015 (morphology, pathogenicity and phylogeny).
Table Curvularia. Details of the isolates used in the phylogenetic tree
| Species | Isolate | ITS | GAPDH | TEF1– α |
| Curvularia aeria | CBS 294.61* | HE861850 | HF565450 | |
| C. affinis | CBS 154.34* | KJ909780 | KM230401 | KM196566 |
| C. akaii | CBS 317.86 | KJ909782 | KM230402 | KM196569 |
| C. akaiiensis | BRIP 16080* | KJ415539 | KJ415407 | KJ415453 |
| C. alcornii | MFLUCC 10-0703* | JX256420 | JX276433 | JX266589 |
| C. americana | UTHSC 08-3414* | HE861833 | HF565488 | |
| C. asiatica | MFLUCC 10-0711* | JX256424 | JX276436 | JX266593 |
| C. australiensis | BRIP 12044* | KJ415540 | KJ415406 | KJ415452 |
| C. australis | BRIP 12521* | KJ415541 | KJ415405 | KJ415451 |
| C. bannonii | BRIP 16732* | KJ415542 | KJ415404 | KJ415450 |
| C. borreriae | CBS 859.73 | HE861848 | HF565455 | |
| C. bothriochloae | BRIP 12522* | KJ415543 | KJ415403 | KJ415449 |
| C. brachyspora | CBS 186.50 | KJ922372 | KM061784 | KM230405 |
| C. buchloes | CBS 246.49* | KJ909765 | KM061789 | KM196588 |
| C. carica-papayae | CBS 135941* | HG778984 | HG779146 | |
| C. chiangmaiensis | CPC 28829* | MF490814 | MF490836 | MF490857 |
| C. chlamydospora | UTHSC 07-2764* | HG779021 | HG779151 | |
| C. clavata | BRIP 61680b | KU552205 | KU552167 | KU552159 |
| C. coicis | CBS 192.29* | JN192373 | JN600962 | JN601006 |
| C. crustacea | BRIP 13524* | KJ415544 | KJ415402 | KJ415448 |
| C. cymbopogonis | CBS 419.78 | HG778985 | HG779129 | HG779163 |
| C. dactyloctenicola | CPC 28810* | MF490815 | MF490837 | MF490858 |
| C. dactyloctenii | BRIP 12846* | KJ415545 | KJ415401 | KJ415447 |
| C. ellisii | CBS 193.62* | JN192375 | JN600963 | JN601007 |
| C. eragrostidis | CBS 189.48 | HG778986 | HG779154 | HG779164 |
| C. geniculata | CBS 187.50 | KJ909781 | KM083609 | KM230410 |
| C. gladioli | CBS 210.79 | HG778987 | HG779123 | |
| C. graminicola | BRIP 23186* | JN192376 | JN600964 | JN601008 |
| C. gudauskasii | DAOM 165085 | AF071338 | ||
| C. harveyi | BRIP 57412* | KJ415546 | KJ415400 | KJ415446 |
| C. hawaiiensis | BRIP 11987* | KJ415547 | KJ415399 | KJ415445 |
| C. heteropogonicola | BRIP 14579* | KJ415548 | KJ415398 | KJ415444 |
| C. heteropogonis | CBS 284.91* | JN192379 | JN600969 | JN601013 |
| C. hominis | CBS 136985* | HG779011 | HG779106 | |
| C. homomorpha | CBS 156.60* | JN192380 | JN600970 | JN601014 |
| C. inaequalis | CBS 102.42* | KJ922375 | KM061787 | KM196574 |
| C. intermedia | CBS 334.64 | HG778991 | HG779155 | HG779169 |
| C. ischaemi | CBS 630.82* | JX256428 | JX276440 | |
| C. kusanoi | CBS 137.29 | JN192381 | JN601016 | |
| C. lunata | CBS 730.96* | JX256429 | JX276441 | JX266596 |
| C. malina | CBS 131274* | JF812154 | KP153179 | KR493095 |
| C. miyakei | CBS 197.29* | KJ909770 | KM083611 | KM196568 |
| C. muehlenbeckiae | CBS 144.63* | HG779002 | HG779108 | |
| C. neergaardii | BRIP 12919* | KJ415550 | KJ415397 | KJ415443 |
| C. neoindica | BRIP 17439 | AF081449 | AF081406 | |
| C. nicotiae | CBS 655.74* = BRIP 11983 | KJ415551 | KJ415396 | KJ415442 |
| C. nodosa | CPC 28801 | MF490817 | MF490839 | MF490860 |
| C. nodosa | CPC 28812 | MF490818 | MF490840 | MF490861 |
| C. nodosa | CPC 28800* | MF490816 | MF490838 | MF490859 |
| C. nodulosa | CBS 160.58 | JN601033 | JN600975 | JN601019 |
| C. oryzae | CBS 169.53* | KP400650 | KP645344 | KM196590 |
| C. ovariicola | CBS 470.90* | JN192384 | JN600976 | JN601020 |
| C. pallescens | CBS 156.35* | KJ922380 | KM083606 | KM196570 |
| C. palmicola | MFLUCC 14-0404* | MF621582 | ||
| C. papendorfii | CBS 308.67* | KJ909774 | KM083617 | KM196594 |
| C. perotidis | CBS 350.90* | JN192385 | KJ415394 | JN601021 |
| C. pisi | CBS 190.48* | KY905678 | KY905690 | KY905697 |
| C. portulacae | CBS 239.48* = BRIP 14541 | KJ415553 | KJ415393 | KJ415440 |
| C. prasadii | CBS 143.64* | KJ922373 | KM061785 | KM230408 |
| C. protuberata | CBS 376.65* | KJ922376 | KM083605 | KM196576 |
| C. pseudobrachyspora | CPC 28808* | MF490819 | MF490841 | MF490862 |
| C. pseudolunata | UTHSC 09-2092* | HE861842 | HF565459 | |
| C. pseudorobusta | UTHSC 08-3458 | HE861838 | HF565476 | |
| C. ravenelii | BRIP 13165* | JN192386 | JN600978 | JN601024 |
| C. richardiae | BRIP 4371* | KJ415555 | KJ415391 | KJ415438 |
| C. robusta | CBS 624.68* | KJ909783 | KM083613 | KM196577 |
| C. ryleyi | BRIP 12554* | KJ415556 | KJ415390 | KJ415437 |
| C. senegalensis | CBS 149.71 | HG779001 | HG779128 | |
| C. sesuvi | Bp-Zj 01 | EF175940 | ||
| C. soli | CBS 222.96* | KY905679 | KY905691 | KY905698 |
| C. sorghina | BRIP 15900* | KJ415558 | KJ415388 | KJ415435 |
| C. spicifera | CBS 274.52 | JN192387 | JN600979 | JN601023 |
| C. subpapendorfii | CBS 656.74* | KJ909777 | KM061791 | KM196585 |
| C. trifolii | CBS 173.55 | HG779023 | HG779124 | |
| C. tripogonis | BRIP 12375* | JN192388 | JN600980 | JN601025 |
| C. tropicalis | BRIP 14834* | KJ415559 | KJ415387 | KJ415434 |
| C. tsudae | ATCC 44764* | KC424596 | KC747745 | KC503940 |
| C. tuberculata | CBS 146.63* | JX256433 | JX276445 | JX266599 |
| C. uncinata | CBS 221.52* | HG779024 | HG779134 | |
| C. variabilis | CPC 28815* | MF490822 | MF490844 | MF490865 |
| C. variabilis | CPC 28813 | MF490820 | MF490842 | MF490863 |
| C. variabilis | CPC 28814 | MF490821 | MF490843 | MF490864 |
| C. variabilis | CPC 28816 | MF490823 | MF490845 | MF490866 |
| C. verruciformis | CBS 537.75 | HG779026 | HG779133 | HG779211 |
| C. verruculosa | CBS 150.63 | KP400652 | KP645346 | KP735695 |
| C. verruculosa | CPC 28792 | MF490825 | MF490847 | MF490868 |
| C. verruculosa | CPC 28809 | MF490824 | MF490846 | MF490867 |
| Bipolaris maydis | CBS 137271* | AF071325 | KM034846 | KM093794 |
| B. oryzae | MFLUCC 10-0715* | JX256416 | JX276430 | JX266585 |
Figure Phylogenetic tree generated by maximum likelihood analysis of combined ITS, GAPDH and TEF1- α sequence data of Curvularia species. Related sequences were obtained from GenBank. Eighty-nine strains are included in the analyses, which comprise 1996 characters including gaps. The tree was rooted with Bipolaris maydis (CBS 137271), B. microlaenae (CBS 280.91) and B. oryzae (MFLUCC 10-0715). The best scoring RAxML tree with a final likelihood value of -13720.654431 is presented. The matrix had 716 distinct alignment patterns, with 19.91% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.23274, C = 0.300605, G = 0.240654, T = 0.226; substitution rates AC = 0.761255, AG = 2.478832, AT = 0.794435, CG = 0.897227, CT = 5.171218, GT = 1.000000; gamma distribution shape parameter α = 0.790494. RAxML bootstrap support values ≥ 60% (BT) are shown respectively near the nodes. The scale bar indicates 0.02 changes. T, ET, IsoT, IsoLT, IsoPT, LT and NT indicate ex-type, ex-epitype, ex-isotype, ex-isolectotype, ex-isoparatype, ex-lectotype and ex-neotype strains, respectively.


No Comments