27 Oct Pseudoseptoria
Pseudoseptoria Speg., Anal. Mus. nac. B. Aires, Ser. 3 13: 388 (1910) [1911]
Background
Spegazzini (1910) introduced Pseudoseptoria as an asexual genus typified with Pseudoseptoria donacicola. Wijayawardene et al. (2012) placed the genus under Ascomycota, genera incertae sedis. Quaedvlieg et al. (2013) placed the genus in Dothioraceae and this was accepted by Thambugala et al. (2014). With LSU sequence data, Crous et al. (2017) placed Pseudoseptoria to Saccotheciaceae. Wijayawardene et al. (2017a, b, 2018, 2020) accepted this placement. Species of Pseudoseptoria are recorded as pathogens on Poaceae (Quaedvlieg et al. 2013), impairing the photosynthetic process resulting in yield loss.
Classification – Ascomycota, Pezizomycotina, Dothideomycetes, Dothideomycetidae, Dothideales, Saccotheciaceae
Type species – Pseudoseptoria donacicola Speg.
Distribution – Australia, Canada, India, Italy, New Zealand, Poland, Russia, UK and USA (Dennis1986; Ginns 1986; French 1989; Pennycook 1989; Merezhko 1991; Cunnington 2003; Mulenko et al. 2008; Kamal 2010; Farr and Rossman 2020).
Disease symptoms – halo spot, leaf blotch and stem speckle
Halo spot: Elliptical, tan to brownish-grey spots (<10mm long) with a dark border surrounded by a prominent yellow halo that can be observed on the leaf blade, sometimes covering the entire leaf blade. In older lesions, small pycnidia may be visible (Slopek and Labun 1992; Carmona et al. 1996; Murray et al. 2013).
Leaf blotch: Brown flecks and frog-eye spots on leaf blades can be observed in early spring, which enlarges to straw-coloured blotches scattered with minute pycnidia. These spots may drop out, leaving holes (Horst 2013).
Stem speckle: The disease occurs in the leaves, sheaths, culms, and head spikes. The lesions are rectangular, ash white, (1-2 mm long) with a brown, thin border. The lesion is delimited by leaf veins and becomes distinct with a clear boundary. The conidia formed on the lesions disperse by wind and rain.
Pathogen biology, disease cycle and epidemiology
The pathogen is dispersed through spores in rain splash. Infection requires an extended period of wetness. Spore germination and infection occur optimally at temperatures between 15 ºC and 25 ºC. Spores produced in overwintering crop debris serve as sources of primary inocula (Sinclair and Dhingra1995). Further studies are needed regarding the disease mechanisms and disease cycle.
Hosts – members of Poaceae are susceptible: Alopecurus pratensis, Arrhenatheru melatius, Arundo donax, Bromus species, Dactylis glomerata, Danthonia spicata, Elymus alaskanus, Festu carubra, Hordeum vulgare, Panicum virgatum, Phleum species, Phragmites australis and Poa species (Ginns 1986; Pennycook 1989; Shivas1989; Greuter at al. 1991; Merezhko 1991; Roane et al. 1996; Gravert et al 2002; Mulenko et al. 2008; Farr and Rossman 2020)
Morphological based identification and diversity
The genus is characterized by immersed, branched, septate, pale brown mycelium, pycnidial, solitary or linearly aggregated, immersed, brown, globose, unilocular, thin-walled conidiomata of walls of pale brown cells of textura angularis with distinct, central, circular ostioles. Conidiogenous cells are discrete, determinate or indeterminate, hyaline, smooth, ampulliform with a prominent cylindrical papilla and falcate. Conidia are fusoid, hyaline, aseptate, guttulate, smooth and thin-walled, and acutely rounded at each end (Sutton 1980; Quaedvlieg et al. 2013).
Molecular based identification and diversity
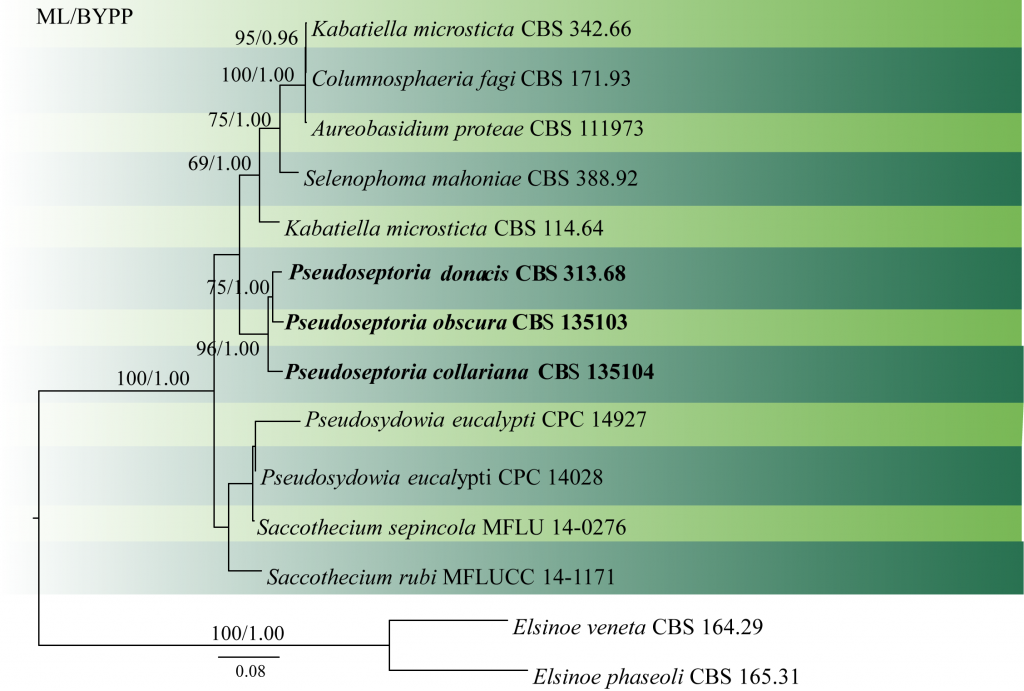
Quaedvlieg et al. (2013) revised the Septoria and septoria-like genera based on morphology and multi loci analyses and introduced two new species. Phylogenetic analysis conducted by Crous et al. (2017) was based only on LSU sequence data. In our analysis, we used LSU, ITS and rpb2 and obtained the same topology (Fig 1).
Recommended genetic marker (genus level) – LSU
Recommended genetic markers (species level) – LSU, ITS and rpb2
Accepted number of species – There are eight epithets listed in Index Fungorum (2020). However, only three species have DNA sequence data (P. collariana, P. donacis and P. obscura) (Table 1).
References – Sutton 1980 (morphology); Quaedvlieg et al. 2013, Crous et al. 2017 (morphology and phylogeny)
Table 1 DNA barcodes available for Pseudoseptoria. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species | Isolate | LSU | ITS | rpb2 |
| Pseudoseptoria collariana | CBS 135104* | KF251721 | KF251218 | KF252223 |
| P. donacis# | CBS 313.68 | MH870852 | MH859141 | |
| P. obscura | CBS 135103 | KF251722 | KF251219 | KF252224 |
Fig. 1 Phylogenetic tree generated by maximum likelihood analysis of combined LSU, ITS and rpb2 sequence data. Fourteen strains are included in the analyses, which comprised 2207 characters including gaps. The tree was rooted with Elsinoe veneta (CBS 164.29) and Elsinoe phaseoli (CBS165.31). Tree topology of the ML analysis was similar to the BYPP analysis. The best scoring RAxML tree with a final likelihood value of -7330.368152 is presented. The matrix had 500 distinct alignment patterns, with 29.98% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.250948, C = 0.235140, G = 0.278289, T = 0.235623; substitution rates AC = 2.242594, AG = 3.171649, AT = 1.699638, CG = 1.459509, CT = 8.754890, GT = 1.000000; gamma distribution shape parameter α = 0.601361 ML bootstrap values ≥65% and BYPP ≥0.90 are shown respectively near the nodes.

No Comments