17 Sep Neofabraea
Neofabraea H.S. Jacks., Rep. Oregon Exp. Sta.: 187 (1913) [1911-1912]
For synonyms see Index Fungorum (2018)
Background
Neofabraea was introduced by Jackson (1913) and typified by N. malicorticis. Neofabraea alba, N. kienholzii, N. malicorticis and N. perennans are pathogens, saprobes or endophytes mostly associated with fruits. Neofabraea are known as the causal agent of bull’s eye rot of apple and pear fruit, which is an important postharvest disease in the Pacific Northwest of the USA, and also occurs in Australia, Canada, Chile, Europe and New Zealand (de Jong et al. 2001; Cunnington 2004; Henriquez et al. 2004; Gariepy et al. 2005; Henriquez 2005; Johnston et al. 2005; Spotts et al. 2009; Soto-Alvear et al. 2013). The Neofabraea complex also cause anthracnose canker and perennial canker on pome trees (Verkley 1999; de jong et al. 2001; Henriquez et al. 2006), canker on Populus sp. (Thompson 1939; Roll-Hansen and Roll-Hansen 1969; Kasanen et al. 2002), coin canker of ash (Rossman et al. 2002), fruit rot on kiwifruit (Johnston et al. 2004), fruit spot on olive (Rooney-Latham et al. 2013), and leaf spot on citrus (Zhu et al. 2012).
Classification – Leotiomycetes, Leotiomycetidae, Helotiales, Dermataceae
Type species – Neofabrea malicorticis (Cordley) H.S. Jacks., Rep. Oregon Exp. Sta.: 187 (1913) [1911-1912]
Distribution – Worldwide
Disease Symptoms – Anthracnose and perennial canker, Bulls’ eye rot, Fruit rot
Bulls’ eye rot (mainly caused by N. alba, N. kienholzii and N. perennans) lesion is circular, flat to slightly sunken and appears light brown to dark brown with a light coloured centre on fruits (Spotts et al. 2009). Anthracnose cankers caused by N. alba, N. malicorticis and N. perennans appear as small circular spots that are reddish when moist. These lesions become elongated and sunken as they enlarge and orange to brown, with cracks around the edges. As damaged bark disintegrates, the canker develops a “fiddle string” appearance. Perennial canker is very similar to the young anthracnose canker. Sunken, elliptical, discoloured areas in the bark can be observed. As the cankers age, formation of callus tissue will result in a series of concentric rings (Henriquez et al. 2006).
Hosts – Actinidia sp., Aucuba sp., Chamaecyparis sp., Ilex sp., Malus sp., Olea sp., Populus sp., Pyrus sp.
Morphological based identification and diversity
Neofabraea was introduced based on Neofabraea malicorticis (Jackson 1913). This genus is very similar to Pezicula and Nannfeldt (1932) combined the type species N. malicorticis into Pezicula. Some other Neofabraea species were transferred to Pezicula (Seaver 1951; Dugan et al. 1993). With new morphological information and phylogenetic analyses, Neofabraea and Pezicula species were retained in separate genera (Verkley 1999; Abeln et al. 2000), but Pezicula alba resembles Neofabraea alba and was hence synonymised to Neofabraea alba (Verkley 1999). In previous studies, the asexual morphs of Neofabraea have been reported to be Cryptosporiopsis with aseptate, fusiform conidia (and later often septate) (Verkley 1999; Johnston et al 2004; Zhu et al. 2012). To avoid dual nomenclature, species of Cryptosporiopsis have been transferred to Neofabraea. To protect Neofabraea over Phlyctema, a suggestion was made to combine Neofabraea vagabunda under N. alba (Johnston et al. 2014). In the past, the type of species of Neofabrae was confused with Neofabraea perennas. In North America, these two were considered as different species but in Europe, they were considered as the same. Based on multigene phylogenetic analyses, de Jong et al. (2001) provided data to prove that these two taxa were different by vegetative compatibility, canker symptoms and response to chemical treatments.
Usually, the apothecia of Neofabraea and Pezicula are similar, but excipular tissues in Pezicula are less different from Neofabraea (Verkley 1999) and macroconidia of Neofabraea are more strongly curved, but the basal scar is smaller than Pezicula. In Pezicula, there are two types of conidiogenous cells, determinate and phialidic or indeterminate and proliferating percurrently, but in Neofabraea only phialidic conidiogenous cells are found (Chen et al. 2016b). These characters can be used in differentiating these two genera. However, as morphological variation among the species of Neofabrea is limited, identification of species based solely on morphological characters are not encouraged.
Molecular based identification and diversity
Neofabraea perennans was described and moved to Pezicula by Dugan et al. (1993). Multigene phylogenetic analyses (ITS nuclear rDNA, SSU mitochondrial rDNA, TUB2) indicated that Neofabraea can be separated from Pezicula (de Jong et al. 2001). Moreover, the TUB2 gene phylogenies showed that apple pathogens contain four clades with strong support, i.e., N. alba, N. krawtzewii, N. malicorticis and N. perennans (de Jong et al. 2001). Although Index Fungorum (2018) lists 14 species in this genus, only nine species, N. actinidiae, N. alba, N. brailiensis, N. inaequalis, N. kienholzii, N. krawtzewii, N. malicorticis, N. perennans and N. vagabunda have sequence data. Therefore, fresh collections and sequence data are needed for the other species. This study reconstructs the phylogeny of Neofabrea based on analyses of a combined ITS, LSU, RPB2 and TUB2 sequence data. The phylogenetic tree obtained corresponds to previous studies (Chen et al. 2016b; de Jong et al. 2001).
Recommended genetic marker (genus level) – LSU
Recommended genetic marker (species level) – TUB2
TUB2 gene is the best single genetic marker for the genus Neofabraea but combined ITS, LSU, RPB2 and TUB2 sequence data can resolve almost all species of Neofabraea (Chen et al. 2016b).
Accepted number of species: There are 14 species epithets in Index Fungorum (2018) under this genus. However, only nine species with molecular data are accepted.
References: Verkley 1999 (morphology and pathogenicity), de Jong et al. 2001, Chen et al. 2016b (phylogeny), Wang et al. 2015 (morphology and a key to species)
Table Neofabraea. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher stains are bolded
| Species | Isolate | ITS | LSU | RPB2 | Btub | |||
| Neofabraea actinidiae | CBS 194.69 | – | KR858871 | KR859320 | KR859286 | |||
| N. actinidiae | CBS 121403 | – | KR858870 | KR859319 | KR859285 | |||
| N. alba | ATCC 38338 | AF281366 | – | – | AF281456 | |||
| N. alba | CBS 452.64 | – | – | – | AF281457 | |||
| N. brasiliensis | CNPUV499* | KR107002 | – | – | KR107011 | |||
| N. brasiliensis | CNPUV506 | KR107001 | – | – | KR107010 | |||
| N. inaequalis | CBS 326.75 | KR859081 | KR858872 | KR859321 | KR859287 | |||
| N. kienholzii | CBS 126461 | KR859082 | KR858873 | KR859322 | KR859288 | |||
| N. kienholzii | CBS 318.77 | KR859083 | KR858874 | KR859323 | KR859289 | |||
| N. krawtzewii | CBS 102867 | KR859084 | KR858875 | KR859324 | AF281459 | |||
| N. krawtzewii | CBS 102868 | – | – | – | KR866108 | |||
| N. malicorticis | CBS 102863 | KR859085 | KR858876 | KR859325 | KR859290 | |||
| N. malicorticis | CBS 122030 | NR144926 | KR858877 | KR859326 | KR859291 | |||
| N. perennans | CBS 102869 | KR859087 | KR858878 | KR859327 | KR866100 | |||
| N. perennans | CBS 275.29 | KR859088 | KR858879 | KR859328 | KR859292 | |||
| N. perennans | CBS 453.64 | KR859089 | KR858880 | KR859329 | KR866102 | |||
| N. vagabunda | M888 | – | – | – | KT963932 | |||
| Pezicula carpinea | CBS 923.96 | KR859108 | KR858899 | KF376158 | KF376279 | |||
| P. carpinea | CBS 921.96 | KR859107 | KR858898 | KF376159 | KF376278 | |||
| P. acericola | CBS 239.97 | KR859093 | KR858884 | KF376214 | KF376283 | |||
| P. brunnea | CBS 120291 | KR859103 | KR858894 | – | – | |||
| P. aurantiaca | CBS 201.46 | KR859102 | KR858893 | KF376210 | KF376335 | |||
| Dermea cerasi | KUS-F50981* | – | JN086690 | – | – | |||
| Parafabraea eucalypti | CBS 124810 | KR859091 | GQ303310 | KR859331 | KR859294 | |||
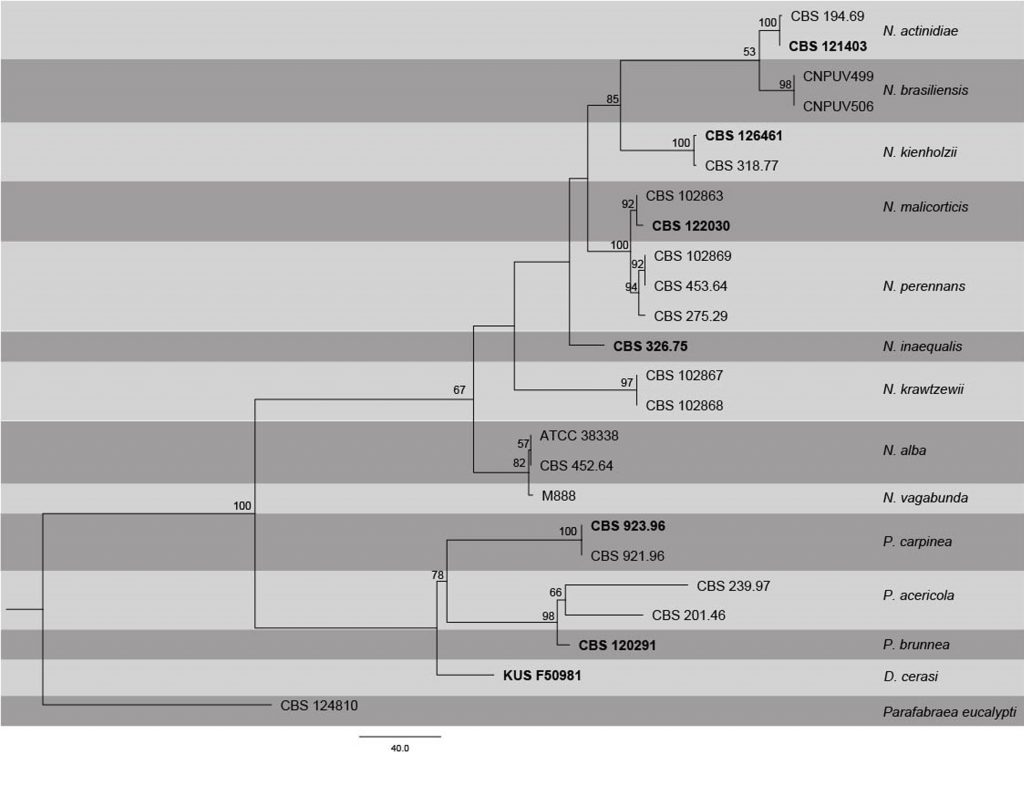
Fig. Phylogenetic tree generated by maximum Parsimony analysis of combined ITS, LSU, RPB2 and TUB2 sequence data of Neofabraea species. Related sequences were obtained from GenBank. Twenty four strains are included in the analyses, which comprise 2767 characters including gaps. Single gene analyses were carried out (not shown) and the phylogeny generated were the same as combined analyses. The tree was rooted with Parafabraea eucalypti (CBS 124810). The maximum parsimonious dataset consisted of constant 2153, 463 parsimony-informative and 151 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 1023 steps (CI = 0.742, RI 0.825, RC = 0.612, HI = 0.258) in the first tree. Maximum parsimony bootstrap support values ≥50% (BT) are shown respectively near the nodes. The scale bar indicates 40.0 changes. The ex-type strains are in bold.

No Comments