24 Oct Cylindrocladiella
Cylindrocladiella Boesew., Canadian Journal of Botany 60 (11): 2289 (1982)
= Nectricladiella Crous & C.L. Schoch, Studies in Mycology 45: 54 (2000)
Background
Boeswinkel (1982) established Cylindrocladiella to accommodate five Cylindrocladium-like species producing small, cylindrical conidia. Even though the generic status of Cylindrocladiella was initially opposed by Crous and Wingfield (1993), later studies on morphological comparisons by Crous et al. (1994) and molecular data (Victor et al. 1998; Schoch et al. 2000) supported the establishment of Cylindrocladiella as a genus. This genus is commonly confused with the asexual morph of Calonectria but can be distinguished by clear morphological differences, such as aseptate stipe extensions, different branching patterns of the conidiophores and comparatively small, aseptate conidia. Although species are generally not regarded as important plant pathogens, correct identification is essential for disease control and biosecurity implications.
Classification – Ascomycota, Sordariomycetes, Hypocreomycetidae, Hypocreales, Nectriaceae
Type species – Cylindrocladiella parva (P.J. Anderson) Boesew.
Distribution – as a soil-borne fungus, the species in Cylindrocladiella have a cosmopolitan distribution in various geographically and climatically distinct regions around the world (Farr and Rossman 2020).
Disease symptoms – black-foot disease, damping-off, leaf spot, root rot and shoot die-back
Many species belonging to Cylindrocladiella are opportunistic plant pathogens but they are not considered as primary pathogens. They can be isolated associated with disease symptoms such as leaf spot, damping off and shoot die-back (Scattolin and Montecchio 2007; Pham et al. 2018). Chocolate brown lesions around the shoots spread primarily to be followed by wilting of the shoot tip, reddish discolouration, dropping of leaves, and finally plant death (Brielmaier-Liebetanz et al. 2013). Characteristic symptoms of the black-foot disease include a reduction in root biomass and root hairs with sunken and necrotic root lesions (Agustí-Brisach and Armengol 2013). Symptoms of Cylindrocladiella root rot are black lesions on the tap and lateral roots, wilting and foliar necrosis, and the outer bark of the seedlings will crack and become loose (Sinclair and Lyon 2005).
Hosts — Species are soil-borne, weak pathogens of forestry, agricultural and horticultural crops. There are 270 records of Cylindrocladiella associated with different plant species (Farr and Rossman 2020). Among them, different Vitis species and Eucalyptus species are common hosts associated with different species of Cylindrocladiella.
Morphological based identification and diversity
Cylindrocladiella can be distinguished from related species by penicillate and/or subverticillate symmetrically branched conidiophores which produce small, cylindrical, 1-septate conidia and aseptate stipe extensions (Lombard et al. 2012). The generic status of Cylindrocladiella was earlier strongly contested (Sharma and Mohanan 1991), however, based on morphological evaluation and comparisons by Crous and Wingfield (1993) and Crous et al. (2017) confirmed its generic status. Victor et al. (1998) and Schoch et al. (2000) provided molecular data to support generic status. Lombard et al. (2011) in his revision of Cylindrocladiella mentioned that only two species have been recognized with their respective Nectricladiella sexual morph. Rossman et al. (2013) proposed that the generic name Cylindrocladiella be used rather than Nectricladiella. Lombard et al. (2015) showed that Cylindrocladiella formed a monophyletic group in Nectriaceae (Wijayawardene et al. 2020).
Molecular based identification and diversity
Using RFLPs and AT-DNA data, Victor et al. (1998) recognised seven species in the genus. Schoch et al. (2000) added another species based on ITS and partial tub2. Van Coller et al. (2005) introduced the use of his3 sequence data for this group. A combined multilocus phylogeny of his, tef1, tub2 and ITS was used by Lombard et al. (2011) which resulted in 18 new Cylindrocladiella species and several unresolved species complexes. Lombard et al. (2017) introduced six new species based on a combined ITS, tef1 and tub2 dataset. Pham et al. (2018) introduced five new species based on his, tef1, tub2 and ITS sequence data and Marin-Felix et al. (2019) introduced two new species based on ITS, tef1 and tub2 sequence data. Here we reconstruct the phylogenetic analyses of these species based on ITS, tef1 and tub2 sequence data (Fig. 1).
Recommended genetic markers (genus level) – ITS, LSU
Recommended genetic markers (species level) – his, tef1, tub2
Accepted number of species – There are 47 species epithets in Index Fungorum (2020). However, only 46 species have DNA sequence data (Table 1).
References –Crous and Wingfield 1993, Lombard et al. 2011 (morphology); Victor et al. 1998, Schoch et al. 2000, Lombard et al. 2015 (morphology, phylogeny).
Table 1 DNA barcodes available for Cylindrocladiella. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species | Isolate No | ITS | tef1 | tub2 |
| Cylindrocladiella addiensis | CBS 143794* | MH111383 | MH111393 | MH111388 |
| C. arbusta# | CMW 47295* | MH017015 | MH016977 | MH016958 |
| C. australiensis | CBS 129567* | JN100624 | JN099060 | JN098747 |
| C. brevistipitata | CBS 142786* | – | MF444940 | MF444926 |
| C. camelliae | IMI 346845 | AF220952 | JN099087 | AY793471 |
| C. clavata | CBS 129564* | JN099095 | JN098974 | JN098752 |
| C. cymbiformis | CBS 129553* | JN099103 | JN098988 | JN098753 |
| C. elegans | CBS 338.92* | AY793444 | JN099039 | AY793474 |
| C. ellipsoidea | CBS 129573* | JN099094 | JN098973 | JN098757 |
| C. hahajimaensis | MAFF 238172* | JN687561 | JN687562 | – |
| C. hawaiiensis | CBS 129569* | JN100621 | JN099057 | JN098761 |
| C. horticola | CBS 142784* | MF444911 | MF444938 | MF444924 |
| C. humicola | CBS 142779* | MF444906 | MF444933 | MF444919 |
| C. infestans | CBS 111795* | AF220955 | JN099037 | AF320190 |
| C. kurandica | CBS 129577* | JN100646 | JN099083 | JN098765 |
| C. lageniformis | CBS 340.92* | AF220959 | JN099003 | AY793481 |
| C. lanceolata | CBS 129566* | JN099099 | JN098978 | JN098789 |
| C. lateralis | CBS 142788* | MF444914 | MF444942 | MF444928 |
| C. longiphialidica | CBS 129557* | JN100585 | JN098966 | JN098790 |
| C. longistipitata | CBS 116075* | AF220958 | JN098993 | AY793506 |
| C. malesiana# | CMW 48278*
|
MH017019
|
MH016981
|
MH016962
|
| C. microcylindrica | CBS 111794* | AY793452 | JN099041 | AY793483 |
| C. natalensis | CBS 114943* | JN100588 | JN099016 | JN098794 |
| C. nederlandica | CBS 152.91* | JN100603 | JN099033 | JN098800 |
| C. novazelandica | CBS 486.77* | AF220963 | JN099050 | AY793485 |
| C. nauliensis | CBS 143792* | MH111387 | MH111397 | MH111392 |
| C. obpyriformis# | CMW 47194*
|
MH017022
|
– | MH016966 |
| C. parva | CBS 114524 | AF220964 | JN099009 | AY793486 |
| C. parvispora# | CMW 47197* | MH017025 | MH016987 | MH016968 |
| C. peruvianum | IMUR 1843* | AF220966 | JN098968 | AY793500 |
| C. postalofficium | CPC 37513*
|
MN562148
|
– | MN556845 |
| C. pseudocamelliae | CBS 129555* | JN100577 | JN098958 | JN098814 |
| C. pseudohawaiiensis | CBS 210.94* | JN099128 | JN099012 | JN098819 |
| C. pseudoinfestans | CBS 114531* | AF220957 | JN099004 | AY793508 |
| C. pseudoparva | CBS129560* | JN100620 | JN099056 | JN098824 |
| C. queenslandica | CBS 129574* | JN099098 | JN098977 | JN098826 |
| C. reginae | CBS 142782* | MF444909 | MF444936 | MF444922 |
| C. solicola# | CMW47198* | MH017021 | MH016983 | MH016964 |
| C. stellenboschensis | CBS 110668* | JN100615 | JN099051 | JN098829 |
| C. terrestris | CBS 142789* | MF444915 | MF444943 | MF444929 |
| C. thailandica | CBS 129571* | JN100582 | JN098963 | JN098834 |
| C. variabilis | CBS 129561* | JN100643 | JN099080 | JN098719 |
| C. viticola# | CBS 112897* | AY793468 | JN099064 | AY793504 |
| C. vitis# | CBS 142517* | KY979751 | KY979891 | KY979918 |
| C. xishuangbannaensis | KUMCC 16-0146* | MH388337 | MH388372 | – |
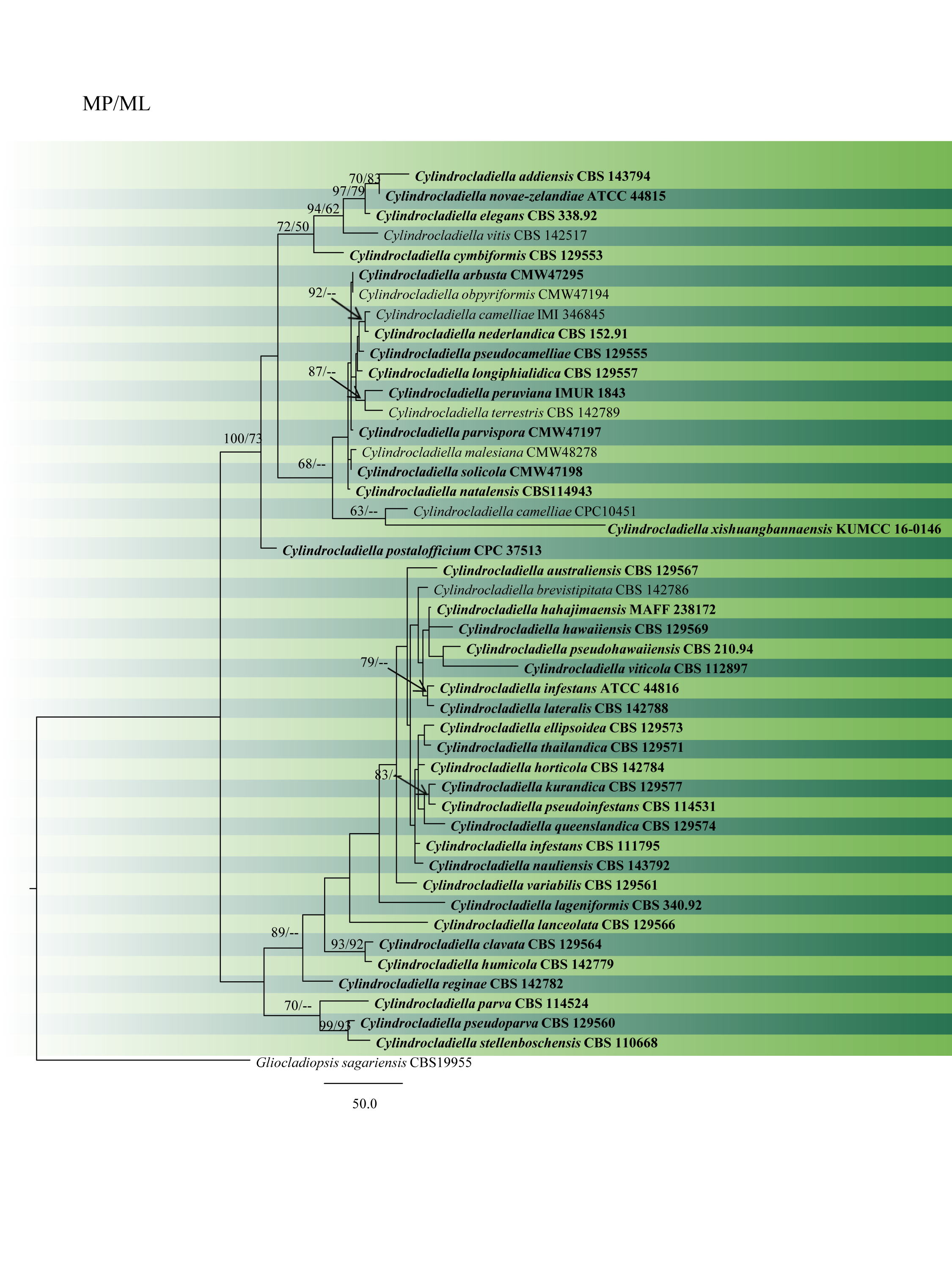 Fig. 1 Phylogram generated from MP analysis based on combined sequences of ITS, tef1 and tub2 sequences of all the accepted species of Cylindrocladiella. Related sequences were obtained from GenBank. Fourty-six taxa are included in the analyses, which comprise 2460 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Gliocladiopsis sagariensis (CBS 19955). The best scoring RAxML tree with a final likelihood value of – 6772.195394 is presented. The matrix had 261 distinct alignment patterns, with 0.96% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.230657, C = 0.279364, G = 0.252128, T = 0.237852; substitution rates AC = 1.388608, AG = 2.845402, AT = 2.389715, CG = 0.838197, CT = 7.220493, GT = 1.000000; gamma distribution shape parameter a = 0.650385. Maximum likelihood and MP bootstrap support value >50% are shown respectively near the nodes. Ex-type strains are in bold.
Fig. 1 Phylogram generated from MP analysis based on combined sequences of ITS, tef1 and tub2 sequences of all the accepted species of Cylindrocladiella. Related sequences were obtained from GenBank. Fourty-six taxa are included in the analyses, which comprise 2460 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Gliocladiopsis sagariensis (CBS 19955). The best scoring RAxML tree with a final likelihood value of – 6772.195394 is presented. The matrix had 261 distinct alignment patterns, with 0.96% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.230657, C = 0.279364, G = 0.252128, T = 0.237852; substitution rates AC = 1.388608, AG = 2.845402, AT = 2.389715, CG = 0.838197, CT = 7.220493, GT = 1.000000; gamma distribution shape parameter a = 0.650385. Maximum likelihood and MP bootstrap support value >50% are shown respectively near the nodes. Ex-type strains are in bold.

No Comments