17 Sep Phellinotus
Phellinotus Drechsler-Santos et al., in Drechsler-Santos et al., Phytotaxa 261(3): 222 (2016)
Background
Phellinotus was described by Drechsler-Santos et al. (2016) and it is typified by Phellinotus neoaridus Drechsler-Santos & Robledo. Only two species, P. neoaridus and P. piptadeniae (Teixeira) Drechsler-Santos & Robledo have been reported, mostly on living members of Fabaceae (Drechsler-Santos et al. 2010, 2016; Salvador-Montoya et al. 2015). Phellinotus is characterized by the annual to perennial, pileate, applanate to ungulate, fulvous brown to dark brown basidiomata; brown to blackened, rugose to rimose pileus; context with a black line near/below the upper surface, distinct or indistinct; and stratified tubes, with or without contextual tissue layer between them. The hymenophore is poroid, with pores irregularly rounded, fulvous brown to deep brown. The hyphal system is dimitic with skeletal hyphae restricted to the trama of tube layer: in the context, generative hyphae thin- to thick-walled, first regularly septate, branched, becoming sclerified and some portions of thick-walled hyphae sparsely simple-septate, and in the trama, simple septate generative and skeletal hyphae. Setae and other sterile elements are absent. The basidiospores are broadly ellipsoid to ellipsoid, adaxially flattened, smooth, thick-walled and yellow in lactophenol, becoming chestnut brown in KOH solution, weakly cyanophilous, IKI- (Drechsler-Santos et al. 2016). Phellinotus neoaridus is very common on Caesalpinia spp., while P. piptadeniae on Piptadeniae spp., with reports also on Libidibia glabrata, Mimosa spp., Pithecellobium excelsum, Senegalia sp. and Eugenia rostrifolia (Salvador-Montoya et al. 2015; Drechsler-Santos et al. 2016).
Classification – Agaricomycetes, incertae sedis, Hymenochaetales, Hymenochataceae
Type species – Phellinotus neoaridus Drechsler-Santos et al., in Drechsler-Santos et al., Phytotaxa 261(3): 222(2016)
Distribution – Brazil, Peru
Disease Symptoms – No evident symptoms in the tree, but when the basidioma is removed, rot is visible; when stems or branches are cut, a black line delimiting a less dense zone in the wood is visible both in the cortex and core. Teixeira (1950) reported defoliation, progressive die-back of twigs and branches, and discoloration of the heartwood of Piptadenia communis older than 8-10 years.
Hosts – mostly on living Fabaceae (Caesalpinia sp., Libidibia glabrata, Mimosa sp., Piptadenia sp., Pithecellobium excelsum, Senegalia sp.), and one report on Myrtaceae (Eugenia rostrifolia).
Morphological based identification and diversity
The two species of Phellinotus were previously identified as Phellinus rimosus (Berk.) Pilát (= Phellinotus neoaridus) and Phellinus piptadeniae Teixeira (= Phellinotus piptadeniae) (Teixeira 1950; Drechsler-Santos et al. 2010). However, after molecular analyses followed by detailed morphological studies, they were accommodated in the new genus Phellinotus and one new species was described (Drechsler-Santos et al. 2016). The geographical distribution of the species is of interest as the type specimen of Phellinus rimosus was from Australia (Tasmania), thus specimens collected elsewhere and identified as such should be re-examined and a new species may be discovered.
This genus can be identified by the morphology of its basidiomata and by the occurrence mostly on living Fabaceae.
Molecular based identification and diversity
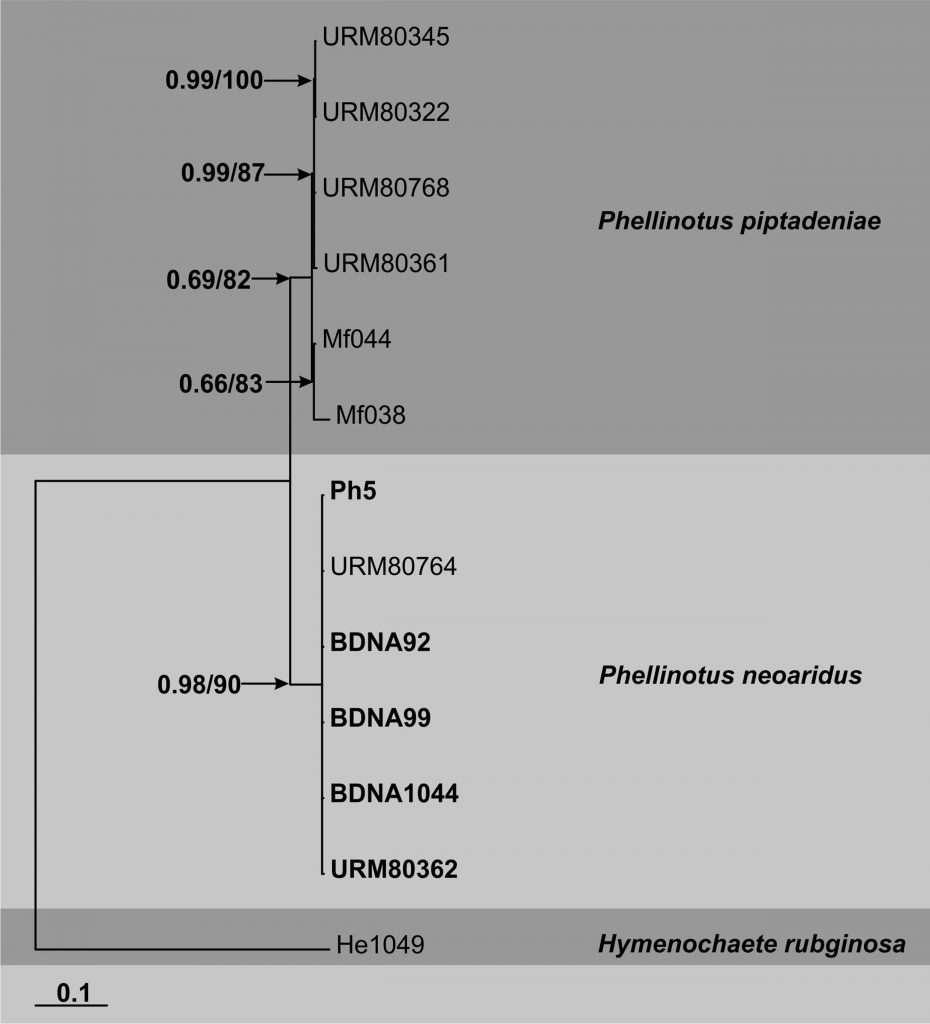
The taxonomy of Phellinotus was recently determined based on a combined dataset of ITS and LSU rDNA sequence data (Drechsler-Santos et al. 2016) of species previously identified as Phellinus rimosus and Phellinus piptadeniae. However, they nested in Fomitiporella (Dai, pers. com.; Crous et al. 2018) and the use of more markers is desirable for a better understanding of their status. Here we present an updated phylogeny for Phellinotus based on the combined analyses of ITS and LSU rDNA sequence data. This tree includes the sequence of the type species of the genus and new sequences of Phellinotus neoaridus.
Recommended genetic marker (genus level) – LSU
Recommended genetic markers (species level) –ITS, TEF1-α and RPB2 as additional markers
Accepted number of species: Two species
References: Teixeira 1950 (morphology), Drechsler-Santos et al. 2010, Salvador-Montoya et al. 2015 (ecology, morphology), Drechsler-Santos et al. (2016) (morphology, phylogeny).
Table Phellinotus. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an *.
| Species | Voucher | LSU | ITS |
| P. neoaridus | URM80764 | KM211287 | – |
| P. neoaridus | PH5 | MG806098 | – |
| P. neoaridus | URM80362* | KM211286 | KM211294 |
| P. neoaridus | URM82501/BDNA1044 | MH048088 | – |
| P. neoaridus | URM84716/BDNA99 | MH048090 | – |
| P. neoaridus | URM85669/BDNA92 | MH048089 | – |
| P. piptadeniae | MF044 | KP412282 | KP412305 |
| P. piptadeniae | MF038 | KP412278 | KP412299 |
| P. piptadeniae | URM80361 | KM211280 | KM211288 |
| P. piptadeniae | URM80345 | KM211283 | KM211291 |
| P. piptadeniae | URM80322 | KM211282 | KM211290 |
| P. piptadeniae | URM80768 | KM211281 | KM211289 |
Fig. Phylogenetic tree generated by Bayesian inference (BI) of combined ITS and LSU rDNA sequence data of Phellinotus species. Thirteen samples are included in the analyses, which comprise 1491 characters including gaps. Tree was rooted with Hymenochaete rubiginosa (He1049). Tree topology of the BI was similar to the maximum likelihood (ML) analysis. The matrix had 633 phylogenetic informative sites (42, 45%). Estimated base frequencies were as follows; A = 0.233, C = 0.222, G = 0.308, T = 0.237; substitution rates AC = 1.000, AG = 1.867, AT = 1.000, CG = 1.000, CT = 1.867, GT = 1.000; gamma distribution shape parameter α = 0.560. Bayesian posterior probabilities and ML bootstrap values ≥50% are shown respectively near the nodes. The scale bar indicates 0.1 changes. Sequences generated in this study and of the types are in bold.

No Comments