27 Oct Phytophthora
Phytophthora de Bary, J. Roy. Agric. Soc. England, ser. 2 12: 240 (1876)
Background
Phytophthora is classified in the kingdom Straminipila within the diploid, alga-like Oomycetes in the Stramenopile clade of the Kingdom Chromista (Cavalier-Smith 1986; Dick 1995; Yoon et al. 2002; Wijayawardene et al. 2020). Phytophthora consists of about 130 described species with many important plant pathogens. The Oomycota are biologically different from main fungal groups within the Kingdom Fungi (Corliss 1994; Cavalier-Smith 1998). For example, their cell walls are made primarily of cellulose instead of chitin as in most fungi and they cannot synthesize β-hydroxysterols, which is vital for synthesizing hormones that regulate sexual reproduction (Hyde et al. 2014). Another important difference is that oomycetes are diploid throughout their life cycle. One similarity between Phytophthora species and Eumycotan fungi is that they both produce hyphae.
Classification – Oomycota, Peronosporales, Peronosporacae
Type species – Phytophthora infestans (Mont.) de Bary
Distribution – worldwide
Disease symptoms – blight, canker, dieback, root rots and wilt
Species can have a large impact on agriculture (e.g. Phytophthora infestans, potato late blight), arbiculture (e.g. Phytophthora ramorum, sudden oak death) and whole ecosystems (e.g. Phytophthora cinnamomi in Australia). Phytophthora species damage plants by killing the tissues and resulting necrosis can be seen in leaves, stems or roots. Some species can cause multiple symptoms on a single host, or cause different symptoms on different hosts (Jung and Blaschke 1996).
Blight: Initial symptom is the development of a “water-soaked” appearance, which progresses into brown or black irregular-shaped spots or wedge-shaped lesions. These lesions are usually not surrounded by a yellow halo (Babadoost 2004; Pande et al. 2011; Ali et al. 2017).
Canker: A dark discoloured necrotic lesion in the inner bark of a tree can be seen often on the stem or branches. However, generally, cankers are visible once the outer bark is removed. Cankers are often seen with a reddish-brown liquid that oozes through the bark (Davidson et al. 2002; Jung et al. 2018).
Dieback: Death of shoot tips, twigs and branch tips can be observed. The infection progresses towards the main stem accompanied by a loss of foliage (Kuske and Benson 1983; Akilli et al. 2013).
Decline and Death: This is a gradual process that will take place over several years. Plants fail to grow and the canopy becomes thin due to loss of foliage. Then the whole canopy or sections of the canopy may die (Marais 1980; Belisario et al. 2004; González et al. 2020).
Rot: Dark discoloured rotten tissues that are common on roots, but sometimes extend above the soil surface. However, collar rot occurs at the base of the trunk and extends just below the soil line (Jung and Blaschke 1996; Graham et al. 2011; Summerell and Liew 2020).
Wilting: This is the first above-ground symptom of root rot. Foliage becomes flaccid due to lack of water intake (Vettraino et al. 2009; Xiong et al. 2019).
Phytophthora causes disease in important agricultural and ecological plants. Phytophthora infestans was responsible for the Irish potato famine from 1845 to 1852, causing the death of over 1 million people. Phytophthora ramorum has resulted in the death of millions of coast live oak, tanoak and Japanese larch trees, thus altering the forest ecosystems in California and Oregon, USA (Goheen et al. 2002; Rizzo et al. 2002, 2005).
Hosts – Phytophthora agathidicida (commonly known as kauri dieback), which causes kauri death, is considered as one of the world’s most feared fungi (Hyde et al. 2018a). An extensive survey in previously unexplored ecosystems such as natural forests (Rea et al. 2010; Vettraino et al. 2011; Jung et al. 2011, 2017; Reeser et al. 2013), streams (Reeser et al. 2007; Bezuidenhout et al. 2010; Yang et al. 2016; Brazee et al. 2017), riparian ecosystems (Brasier et al. 2003, 2004; Hansen et al. 2012), and irrigation systems (Hong et al. 2010, 2012; Yang et al. 2014a, b) has led an exponential increase in the number of species.
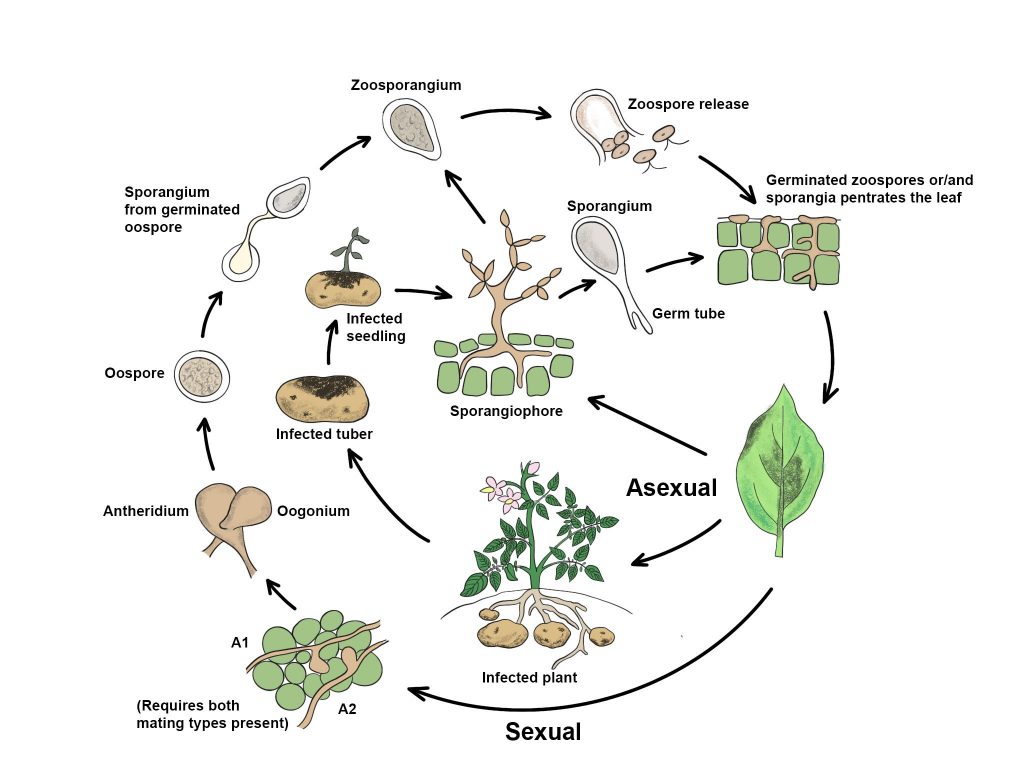
Pathogen biology, disease cycle and epidemiology
Fig. 1 Disease cycle of Phytophthora infestans (redrawn from Bengtsson 2013)
Morphological based identification and diversity
Species-level classification is based on the morphological characterization of reproductive structures including the sporangium (asexual) and oospore (sexual) as well as the production of chlamydospores (Martin et al. 2012). Characteristics that are important for species classification include the diameter of the oogonium and oospore, thickness of the oospore wall, whether or not the oospore fills the oogonium, ornamentation on the oogonial wall, and mode of attachment of the antheridium (Hyde et al. 2014). Identification and classification of Phytophthora species into morphological groups based on several characteristics was initially based on the key provided by Waterhouse (1963), which was later updated by Stamps et al. (1990).
Molecular based identification and diversity
Phytophthora has been historically placed in the Pythiales with Pythium and related genera, however recent phylogenetic analysis with the large (LSU) or small (SSU) rDNA sequences or cox2 gene has indicated a closer relationship with downy mildew and white rusts (Albugo.) in the Peronosporales (Beakes and Sekimoto 2009; Thines et al. 2009). Additional multigene analyses are vital to clarify the relationship between the Peronosporales and Pythium. Early efforts focusing on the phylogenetic relationships in Phytophthora used nuclear-encoded rDNA, primarily the ITS region (Crawford et al. 1996; Cooke and Duncan 1997; Förster et al. 2000). The first comprehensive study was based on the phylogenetic study of the ITS region (Cooke et al. 2000). The study by Kroon et al. (2004) was based on analysis using two nuclear (tef1, tub2) and two mitochondrial (cox1 and nad1) genes. Subsequent phylogenetic analysis was based on sequences of seven nuclear genetic markers (60S ribosomal protein L10, tub2, enolase, heat shock protein90, large subunit rDNA, TigA gene fusion and tef1) which divided the species into 10 well-supported clades (Blair et al. 2008). The phylogenetic study by Martin et al. (2014) was based on seven nuclear and four mitochondrial genes (cox2, nad9, rps10 and secY). More recently, an extensive study of the genus by Yang et al. (2017) was based on sequences of seven nuclear genetic markers as in Blair et al. (2008).
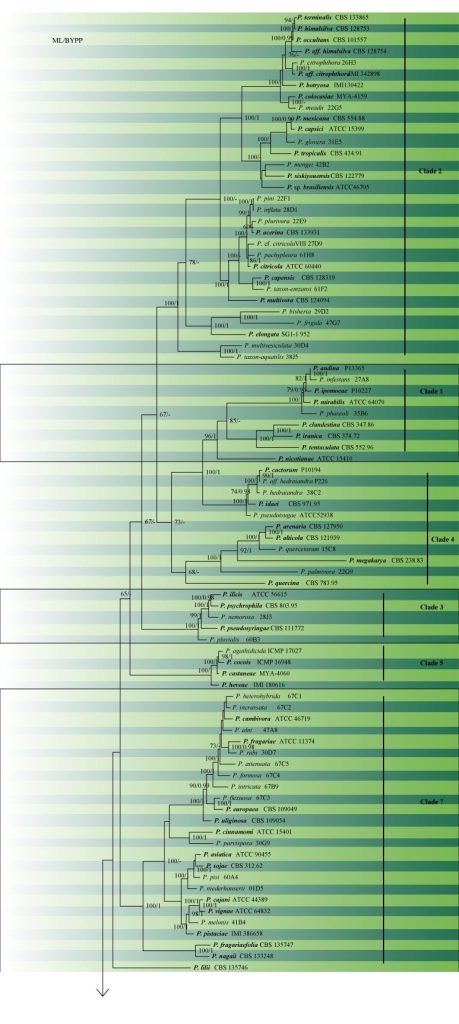
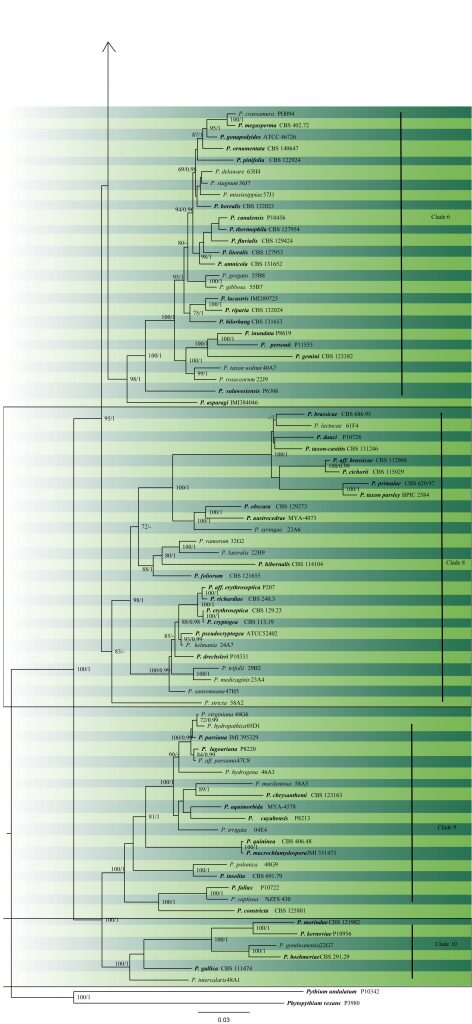
The number of described species in Phytophthora was approximately 55 in 1999, but since then there has been a significant increase in the number of species nearly doubling the number of described species to 105 (Brasier 2007), and over 128 species (Hyde et al. 2014). Additional species have since been described, for example, P. cocois (Weir et al. 2015), P. crassamura (Scanu et al. 2015), P. attenuata, P. xheterohybrida, P. xincrassata (Jung et al. 2017) bringing the total to over 150 species (Jung et al. 2019). The phylogenetic tree constructed is presented in Fig. 2 and the accepted species are given in table 1.
Recommended genetic markers (genus level) – LSU, SSU and cox2
Recommended genetic markers (species level) – LSU, tub2 and cox2
Accepted number of species– There are 317 epithets listed in Index Fungorum (2020), however only 162 species have DNA sequence data (Table 1).
References – Waterhouse 1963, Stamps et al. 1990 (morphology); Crawford et al. 1996, Cooke and Duncan 1997, Cooke et al. 2000, Förster et al. 2000, Brasier 2007, Blair et al. 2008 (morphology and phylogeny); Hyde et al. 2014 (phylogeny and accepted species).
Table 1 DNA barcodes available for Phytophthora. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains and voucher strains are in bold. Species confirmed with pathogenicity studies are marked with #. (VdPN) are strains used by Van der Plaäts-Niterink (1981) for descriptions.
| Species | Strain | 60S | tub2 | tef1 | Enl | HSP90 | 28S | TigA |
| Phytophthora ×alni | 47A8 | KX251595 | KX251596 | KX251597 | KX251598 | KX251599 | KX251600 | KX251601 |
| P. ×heterohybrida | 67C1 | KX251637 | KX251638 | KX251639 | KX251640 | KX251641 | KX251642 | KX251643 |
| P. ×incrassata | 67C2 | KX251644 | KX251645 | KX251646 | KX251647 | KX251648 | KX251649 | KX251650 |
| P. ×stagnum# | 36J7 | KX251368 | KX251369 | KX251370 | KX251371 | KX251372 | KX251373 | KX251374 |
| P. acerina# | 61H1=CBS 133931 | KX250712 | KX250713 | KX250714 | KX250715 | KX250716 | KX250717 | KX250718 |
| P. aff. brassicae# | CBS 112968 | EU079880 | EU079881 | EU079882 | EU079883 | EU079884 | EU079885 | EU079886 |
| P. aff. citrophthora# | IMI 342898 | EU080384 | EU080385 | EU080386 | EU080387 | EU080388 | EU080389 | EU080390 |
| P. aff. erythroseptica | 33A1=P207 | KX251979 | KX251980 | KX251981 | KX251982 | KX251983 | KX251984 | KX251985 |
| P. aff. hedraiandra# | 33F4=P226 | KX250411 | KX250412 | KX250413 | KX250414 | KX250415 | KX250416 | KX250417 |
| P. aff. himalsilva | 61G4=CBS 128754 | KX250621 | KX250622 | KX250623 | KX250624 | KX250625 | KX250626 | KX250627 |
| P. aff. parsiana# | 47C8 | KX252397 | KX252398 | KX252399 | KX252400 | KX252401 | KX252402 | N/A |
| P. agathidicida# | 67D5=ICMP 17027 | KX251076 | KX251077 | KX251078 | KX251079 | KX251080 | KX251081 | KX251082 |
| P. alticola | 47G5=CBS 121939 | KX251006 | KX251007 | KX251008 | KX251009 | KX251010 | KX251011 | KX251012 |
| P. amnicola | 61G6=CBS 131652 | KX251167 | KX251168 | KX251169 | KX251170 | KX251171 | KX251172 | KX251173 |
| P. andina# | P13365 | EU080182 | EU080183 | EU080184 | EU080185 | EU080186 | EU080187 | EU080188 |
| P. aquimorbida | 40A6=MYA–4578 | KX252238 | KX252239 | KX252240 | KX252241 | KX252242 | KX252243 | KX252244 |
| P. arenaria | 55C2=CBS 127950 | KX251013 | KX251014 | KX251015 | KX251016 | KX251017 | KX251018 | KX251019 |
| P. asiatica | 45G1=ATCC 90455 | KX251651 | KX251652 | KX251653 | KX251654 | KX251655 | KX251656 | KX251657 |
| P. asparagi# | 33D7= IMI384046 | KX251466 | KX251467 | KX251468 | KX251469 | KX251470 | KX251471 | KX251472 |
| P. attenuata | 67C5 | KX251609 | KX251610 | KX251611 | KX251612 | KX251613 | KX251614 | KX251615 |
| P. austrocedrae | 41B5=MYA–4073 | KX252161 | KX252162 | KX252163 | KX252164 | KX252165 | KX252166 | KX252167 |
| P. bilorbang# | 61G8=CBS 131653 | KX251181 | KX251182 | N/A | KX251183 | KX251184 | KX251185 | KX251186 |
| P. bisheria | 29D2 | KX250873 | KX250874 | KX250875 | KX250876 | KX250877 | KX250878 | KX250879 |
| P. boehmeriae# | 45F9=CBS 291.29 | EU080161 | EU080162 | EU080163 | EU080164 | EU080165 | EU080166 | EU080167 |
| P. borealis | 60B2=CBS 132023 | KX251187 | KX251188 | KX251189 | KX251190 | KX251191 | KX251192 | KX251193 |
| P. botryosa# | IMI130422=P6945 | EU079934 | EU079935 | EU079936 | EU079937 | EU079938 | EU079939 | EU079940 |
| P. brassicae# | 29D8=CBS 686.95 | KX251993 | KX251994 | KX251995 | KX251996 | KX251997 | KX251998 | KX251999 |
| P. cactorum# | 22E6=P10194 | KX250369 | KX250370 | KX250371 | KX250372 | KX250373 | KX250374 | KX250375 |
| P. cajani# | 45F6=ATCC 44389 | KX251679 | KX251680 | KX251681 | KX251682 | KX251683 | KX251684 | KX251685 |
| P. cambivora# | 22F6=ATCC 46719 | KX251494 | KX251495 | KX251496 | KX251497 | KX251498 | KX251499 | KX251500 |
| P. capensis | 62C1=CBS 128319 | KX250726 | KX250727 | KX250728 | KX250729 | KX250730 | KX250731 | KX250732 |
| P. capsici# | 22F4=ATCC 15399 | KX250635 | KX250636 | KX250637 | KX250638 | KX250639 | KX250640 | KX250641 |
| P. captiosa# | 46H8=NZFS 430 | KX252554 | KX252555 | KX252556 | KX252557 | KX252558 | KX252559 | KX252560 |
| P. castaneae | 22H6=MYA–4060 | KX251083 | KX251084 | KX251085 | KX251086 | KX251087 | KX251088 | KX251089 |
| P. chrysanthemi# | 61F1=CBS 123163 | KX252266 | KX252267 | KX252268 | KX252269 | KX252270 | KX252271 | KX252272 |
| P. cichorii | 62A8=CBS 115029 | KX252007 | KX252008 | KX252009 | KX252010 | KX252011 | KX252012 | KX252013 |
| P. cinnamomi# | 23B2=ATCC 15401 | KX251804 | KX251805 | KX251806 | KX251807 | KX251808 | KX251809 | KX251810 |
| P. citricola# | 33H8=ATCC 60440 | KX250747 | KX250748 | KX250749 | KX250750 | KX250751 | KX250752 | KX250753 |
| P. citrophthora# | 26H3 | KX250551 | KX250552 | KX250553 | KX250554 | KX250555 | KX250556 | KX250557 |
| P. clandestina | 32G1=CBS 347.86 | EU079866 | EU079867 | EU079868 | EU079869 | EU079870 | EU079871 | EU079872 |
| P. cocois | 67D6=ICMP 16948 | KX251104 | KX251105 | KX251106 | KX251107 | KX251108 | KX251109 | KX251110 |
| P. colocasiae# | 22F8=MYA–4159 | KX250558 | KX250559 | KX250560 | KX250561 | KX250562 | KX250563 | KX250564 |
| P. constricta | 55C3=CBS 125801 | KX252561 | KX252562 | KX252563 | KX252564 | KX252565 | KX252566 | KX252567 |
| P. crassamura | 66C9=PH094 | KX251194 | KX251195 | KX251196 | KX251197 | KX251198 | KX251199 | KX251200 |
| P. cryptogea# | 61H9=CBS 113.19 | KX251867 | KX251868 | KX251869 | KX251870 | KX251871 | KX251872 | KX251873 |
| P. dauci | 32E6=P10728 | KX252028 | KX252029 | KX252030 | KX252031 | KX252032 | KX252033 | KX252034 |
| P. drechsleri# | P10331 | EU079506 | EU079507 | EU079508 | EU079509 | EU079510 | EU079511 | EU079512 |
| P. elongata | 33J3=SG1–1 952 | KX250880 | KX250881 | KX250882 | KX250883 | KX250884 | KX250885 | KX250886 |
| P. erythroseptica | 61J2=CBS 129.23 | KX251895 | KX251896 | KX251897 | KX251898 | KX251899 | KX251900 | KX251901 |
| P. europaea# | 62A2=CBS 109049 | KX251522 | KX251523 | KX251524 | KX251525 | KX251526 | KX251527 | KX251528 |
| P. fallax# | 46J2=P10722 | KX252568 | KX252569 | KX252570 | KX252571 | KX252572 | KX252573 | KX252574 |
| P. flexuosa | 67C3 | KX251616 | KX251617 | KX251618 | KX251619 | KX251620 | KX251621 | KX251622 |
| P. fluvialis | 55B6=CBS 129424 | KX251208 | KX251209 | KX251210 | KX251211 | KX251212 | KX251213 | KX251214 |
| P. foliorum# | 49J8=CBS 121655 | KX252112 | KX252113 | KX252114 | KX252115 | KX252116 | KX252117 | KX252118 |
| P. formosa | 67C4 | KX251623 | KX251624 | KX251625 | KX251626 | KX251627 | KX251628 | KX251629 |
| P. fragariae# | 22G6=ATCC 11374 | KX251529 | KX251530 | KX251531 | KX251532 | KX251533 | KX251534 | KX251535 |
| P. fragariaefolia | 61H4=CBS 135747 | KX251853 | KX251854 | KX251855 | KX251856 | KX251857 | KX251858 | KX251859 |
| P. frigida# | 47G7 | KX250908 | KX250909 | KX250910 | KX250911 | KX250912 | KX250913 | KX250914 |
| P. gallica | 50A1=CBS 111474 | KX252589 | KX252590 | KX252591 | KX252592 | KX252593 | KX252594 | KX252595 |
| P. gemini | 46H1=CBS 123382 | KX251125 | KX251126 | KX251127 | KX251128 | KX251129 | KX251130 | KX251131 |
| P. gibbosa | 55B7 | KX251215 | KX251216 | KX251217 | KX251218 | KX251219 | KX251220 | KX251221 |
| P. glovera | 31E5 | KX250642 | KX250643 | KX250644 | KX250645 | KX250646 | KX250647 | KX250648 |
| P. gonapodyides# | 21J5=ATCC 46726 | KX251229 | KX251230 | KX251231 | KX251232 | KX251233 | KX251234 | KX251235 |
| P. gondwanensis | 22G7 | KX252603 | KX252604 | KX252605 | KX252606 | KX252607 | KX252608 | KX252609 |
| P. gregata# | 55B8 | KX251243 | KX251244 | KX251245 | KX251246 | KX251247 | KX251248 | KX251249 |
| P. hedraiandra# | 38C2 | KX250390 | KX250391 | KX250392 | KX250393 | KX250394 | KX250395 | KX250396 |
| P. heveae# | 22J1=IMI 180616 | KX251111 | KX251112 | KX251113 | KX251114 | KX251115 | KX251116 | KX251117 |
| P. hibernalis# | 32F7=CBS 114104 | KX252126 | KX252127 | KX252128 | KX252129 | KX252130 | KX252131 | KX252132 |
| P. himalsilva | 61G3=CBS 128753 | KX250579 | KX250580 | KX250581 | KX250582 | KX250583 | KX250584 | KX250585 |
| P. hydrogena | 46A3 | KX252280 | KX252281 | KX252282 | KX252283 | KX252284 | KX252285 | KX252286 |
| P. hydropathica# | 05D1 | KX252294 | KX252295 | KX252296 | KX252297 | KX252298 | KX252299 | KX252300 |
| P. idaei | 34D4=CBS 971.95 | EU080129 | EU080130 | EU080131 | EU080132 | EU080133 | EU080134 | EU080135 |
| P. ilicis# | 23A7=ATCC 56615 | KX250936 | KX250937 | KX250938 | KX250939 | KX250940 | KX250941 | KX250942 |
| P. infestans# | 27A8 | KX250474 | KX250475 | KX250476 | KX250477 | KX250478 | KX250479 | KX250480 |
| P. inflata# | 28D1 | KX250761 | KX250762 | KX250763 | KX250764 | KX250765 | KX250766 | KX250767 |
| P. insolita# | 38E1=CBS 691.79 | EU080175 | EU080176 | EU080177 | EU080178 | EU080179 | EU080180 | EU080181 |
| P. intercalaris | 48A1 | KX252617 | KX252618 | KX252619 | KX252620 | KX252621 | KX252622 | KX252623 |
| P. intricata | 67B9 | KX251630 | KX251631 | KX251632 | KX251633 | KX251634 | KX251635 | KX251636 |
| P. inundata# | P8619 | EU080202 | EU080203 | EU080204 | EU080205 | EU080206 | EU080207 | EU080208 |
| P. ipomoeae# | 31B6=P10227 | EU080844 | EU080845 | EU080846 | EU080847 | EU080848 | EU080849 | EU080850 |
| P. iranica# | 61J4=CBS 374.72 | KX250439 | KX250440 | KX250441 | KX250442 | KX250443 | KX250444 | KX250445 |
| P. irrigata | 04E4 | KX252308 | KX252309 | KX252310 | KX252311 | KX252312 | KX252313 | KX252314 |
| P. kernoviae | 46C8=P10956 | EU080041 | EU080042 | EU080043 | EU080044 | EU080045 | EU080046 | KX252631 |
| P. lactucae | 61F4 | KX252042 | KX252043 | KX252044 | KX252045 | KX252046 | KX252047 | KX252048 |
| P. lacustris# | IMI389725=P10337 | EU080530 | EU080531 | EU080532 | EU080533 | EU080534 | EU080535 | EU080536 |
| P. lateralis# | 22H9 | KX252133 | KX252134 | KX252135 | KX252136 | KX252137 | KX252138 | KX252139 |
| P. lilii | CBS 135746 | AB856779 | AB856782 | AB856788 | AB856791 | AB856794 | AB856797 | AB856800 |
| P. litoralis | 55B9=CBS 127953 | KX251278 | KX251279 | KX251280 | KX251281 | KX251282 | KX251283 | KX251284 |
| P. macilentosa | 58A5 | KX252329 | KX252330 | KX252331 | KX252332 | KX252333 | KX252334 | KX252335 |
| P. macrochlamydospora# | G231E9=IMI 351473 | EU080658 | EU080659 | EU080660 | N/A | EU080661 | EU080662 | EU080663 |
| P. meadii# | 22G5 | KX250586 | KX250587 | KX250588 | KX250589 | KX250590 | KX250591 | KX250592 |
| P. medicaginis# | 23A4 | KX251902 | KX251903 | KX251904 | KX251905 | KX251906 | KX251907 | KX251908 |
| P. megakarya# | 61J5=CBS 238.83 | KX251034 | KX251035 | KX251036 | KX251037 | KX251038 | KX251039 | KX251040 |
| P. megasperma# | 62C7=CBS 402.72 | KX251285 | KX251286 | KX251287 | KX251288 | KX251289 | KX251290 | N/A |
| P. melonis# | 41B4 | KX251700 | KX251701 | KX251702 | KX251703 | KX251704 | KX251705 | KX251706 |
| P. mengei | 42B2 | KX250656 | KX250657 | KX250658 | KX250659 | KX250660 | KX250661 | KX250662 |
| P. mexicana | 45G4=CBS 554.88 | KX250670 | KX250671 | KX250672 | KX250673 | KX250674 | KX250675 | KX250676 |
| P. mirabilis | 30C2=ATCC 64070 | KX250488 | KX250489 | KX250490 | KX250491 | KX250492 | KX250493 | KX250494 |
| P. mississippiae | 57J1 | KX251291 | KX251292 | KX251293 | KX251294 | KX251295 | KX251296 | KX251297 |
| P. morindae | 62B5=CBS 121982 | KX252633 | KX252634 | KX252635 | KX252636 | KX252637 | KX252638 | KX252639 |
| P. multivesiculata# | 30D4 | KX250922 | KX250923 | KX250924 | KX250925 | KX250926 | KX250927 | KX250928 |
| P. multivora# | 55C5=CBS 124094 | KX250775 | KX250776 | KX250777 | KX250778 | KX250779 | KX250780 | KX250781 |
| P. nagaii | 61H5=CBS 133248 | KX251860 | KX251861 | KX251862 | KX251863 | KX251864 | KX251865 | KX251866 |
| P. nemorosa | 28J3 | KX250957 | KX250958 | KX250959 | KX250960 | KX250961 | KX250962 | KX250963 |
| P. nicotianae# | 22F9=ATCC 15410 | KX250509 | KX250510 | KX250511 | KX250512 | KX250513 | KX250514 | KX250515 |
| P. niederhauserii# | 01D5 | KX251714 | KX251715 | KX251716 | KX251717 | KX251718 | KX251719 | KX251720 |
| P. obscura | 60E9=CBS 129273 | KX252175 | KX252176 | KX252177 | KX252178 | KX252179 | KX252180 | KX252181 |
| P. occultans# | 65B9=CBS 101557 | KX250600 | KX250601 | KX250602 | KX250603 | KX250604 | KX250605 | KX250606 |
| P. ornamentata | 66D2=CBS 140647 | KX251319 | KX251320 | KX251321 | KX251322 | KX251323 | KX251324 | KX251325 |
| P. pachypleura# | 61H8 | KX250796 | KX250797 | KX250798 | KX250799 | KX250800 | KX250801 | KX250802 |
| P. palmivora# | 22G9 | KX251055 | KX251056 | KX251057 | KX251058 | KX251059 | KX251060 | KX251061 |
| P. parsiana# | 47C3=IMI 395329 | KX252357 | KX252358 | KX252359 | KX252360 | KX252361 | KX252362 | KX252363 |
| P. parvispora# | 30G9 | KX251818 | KX251819 | KX251820 | KX251821 | KX251822 | KX251823 | KX251824 |
| P. phaseoli | 35B6 | KX250502 | KX250503 | KX250504 | KX250505 | KX250506 | KX250507 | KX250508 |
| P. pini# | 22F1 | KX250803 | KX250804 | KX250805 | KX250806 | KX250807 | KX250808 | KX250809 |
| P. pinifolia | 47H1=CBS 122924 | KX251333 | KX251334 | KX251335 | KX251336 | KX251337 | KX251338 | KX251339 |
| P. pisi | 60A4 | KX251735 | KX251736 | KX251737 | KX251738 | KX251739 | KX251740 | KX251741 |
| P. pistaciae# | 33D6=IMI 386658 | KX251748 | KX251749 | KX251750 | KX251751 | KX251752 | KX251753 | KX251754 |
| P. plurivora# | 22E9 | KX250817 | KX250818 | KX250819 | KX250820 | KX250821 | KX250822 | KX250823 |
| P. pluvialis# | 60B3 | KX250971 | KX250972 | KX250973 | KX250974 | KX250975 | KX250976 | KX250977 |
| P. polonica | 40G9 | KX252532 | KX252533 | KX252534 | KX252535 | KX252536 | KX252537 | KX252538 |
| P. primulae# | 29E9=CBS 620.97 | KX252063 | KX252064 | KX252065 | KX252066 | KX252067 | KX252068 | KX252069 |
| P. pseudocryptogea# | ATCC52402 | EU080626 | EU080627 | EU080628 | EU080629 | EU080630 | EU080631 | N/A |
| P. pseudosyringae# | 30A8=CBS 111772 | KX250978 | KX250979 | KX250980 | KX250981 | KX250982 | KX250983 | KX250984 |
| P. pseudotsugae | ATCC52938 | EU080426 | EU080427 | EU080428 | EU080429 | EU080430 | EU080431 | EU080432 |
| P. psychrophila | 29J5=CBS 803.95 | KX250992 | KX250993 | KX250994 | KX250995 | KX250996 | KX250997 | KX250998 |
| P. quercetorum | 15C8 | KX251069 | KX251070 | KX251071 | KX251072 | KX251073 | KX251074 | KX251075 |
| P. quercina# | 30A4=CBS 783.95 | KX252647 | KX252648 | KX252649 | KX252650 | KX252651 | KX252652 | KX252653 |
| P. quininea | 45F2=CBS 406.48 | EU080107 | EU080108 | EU080109 | N/A | KX252522 | EU080110 | KX252523 |
| P. ramorum# | 32G2 | KX252147 | KX252148 | KX252149 | KX252150 | KX252151 | KX252152 | KX252153 |
| P. richardiae | 45F5=CBS 240.3 | KX251923 | KX251924 | KX251925 | KX251926 | KX251927 | KX251928 | KX251929 |
| P. riparia | 60B1=CBS 132024 | KX251347 | KX251348 | KX251349 | KX251350 | KX251351 | KX251352 | KX251353 |
| P. rosacearum | 22J9 | KX251431 | KX251432 | KX251433 | KX251434 | KX251435 | KX251436 | KX251437 |
| P. rubi | 30D7 | KX251550 | KX251551 | KX251552 | KX251553 | KX251554 | KX251555 | KX251556 |
| P. sansomeana# | 47H5 | KX251944 | KX251945 | KX251946 | KX251947 | KX251948 | KX251949 | KX251950 |
| P. siskiyouensis# | 41B7=CBS 122779 | KX250677 | KX250678 | KX250679 | KX250680 | KX250681 | KX250682 | KX250683 |
| P. sojae# | 22D8=CBS 312.62 | KX251762 | KX251763 | KX251764 | KX251765 | KX251766 | KX251767 | KX251768 |
| P. sp. brasiliensis | ATCC46705=P0630 | EU080419 | EU080420 | EU080421 | EU080422 | EU080423 | EU080424 | EU080425 |
| P. sp. canalensis | P10456 | EU079569 | EU079570 | EU079571 | EU079572 | EU079573 | EU079574 | N/A |
| P. sp. citricola VIII | 27D9 | KX250838 | KX250839 | KX250840 | KX250841 | KX250842 | KX250843 | KX250844 |
| P. sp. cuyabensis | P8213 | EU080664 | EU080665 | EU080666 | EU080667 | EU080668 | EU080669 | EU080331 |
| P. sp. delaware | 63H4 | KX251396 | KX251397 | KX251398 | KX251399 | KX251400 | KX251401 | KX251402 |
| P. sp. kelmania | 24A7 | KX251986 | KX251987 | KX251988 | KX251989 | KX251990 | KX251991 | KX251992 |
| P. sp. lagoariana | 60B4=P8220 | EU080358 | KX252502 | EU080359 | EU080360 | EU080361 | EU080362 | EU080363 |
| P. sp. personii | P11555 | EU080312 | EU080313 | EU080314 | EU080315 | EU080316 | EU080317 | EU080318 |
| P. sp. sulawesiensis | P6306 | EU080345 | N/A | EU080346 | EU080347 | EU080348 | EU080349 | EU080350 |
| P. stricta | 58A2 | KX252217 | KX252218 | KX252219 | KX252220 | KX252221 | KX252222 | KX252223 |
| P. syringae# | 23A6 | KX252203 | KX252204 | KX252205 | KX252206 | KX252207 | KX252208 | KX252209 |
| P. taxon parsley | 61G1=BPIC 2584 | KX252105 | KX252106 | KX252107 | KX252108 | KX252109 | KX252110 | KX252111 |
| P. taxon walnut | 40A7 | KX251452 | KX251453 | KX251454 | KX251455 | KX251456 | KX251457 | KX251458 |
| P. taxon–aquatilis | 38J5 | KX250929 | KX250930 | KX250931 | KX250932 | KX250933 | KX250934 | KX250935 |
| P. taxon–castitis | 61E7=CBS 131246 | KX252098 | KX252099 | KX252100 | KX252101 | KX252102 | KX252103 | KX252104 |
| P. taxon–emzansi | 61F2 | KX250859 | KX250860 | KX250861 | KX250862 | KX250863 | KX250864 | KX250865 |
| P. tentaculata# | 29F2=CBS 552.96 | EU079955 | EU079956 | EU079957 | EU079958 | EU079959 | EU079960 | EU079961 |
| P. terminalis | 65B8=CBS 133865 | KX250607 | KX250608 | KX250609 | KX250610 | KX250611 | KX250612 | KX250613 |
| P. thermophila | 55C1=CBS 127954 | KX251354 | KX251355 | KX251356 | KX251357 | KX251358 | KX251359 | KX251360 |
| P. trifolii | 29B2 | KX251951 | KX251952 | KX251953 | KX251954 | KX251955 | KX251956 | KX251957 |
| P. tropicalis# | 35C8=CBS 434.91 | KX250698 | KX250699 | KX250700 | KX250701 | KX250702 | KX250703 | KX250704 |
| P. uliginosa | 62A3=CBS 109054 | EU080011 | EU080012 | EU080013 | KX251571 | KX251572 | EU080015 | KX251573 |
| P. vignae# | 45G9=ATCC 64832 | KX251783 | KX251784 | KX251785 | KX251786 | KX251787 | KX251788 | KX251789 |
| P. virginiana | 44G6 | KX252371 | KX252372 | KX252373 | KX252374 | KX252375 | KX252376 | KX252377 |
Fig. 2 Maximum likelihood of Phytophthora based on the concatenated seven nuclear genetic markers (60S Ribosomal protein L10 (60S), beta-tubulin (tub), elongation factor 1 alpha (tef1), enolase (Enl), heat shock protein 90 (hsp90), 28S ribosomal DNA (28S), and tigA gene fusion protein (TigA)). ML bootstrap support values over 60% are indicated and BYPP≥0.90 are shown respectively near the nodes. The type species are in bold. Scale bar indicates number of substitutions per site. The tree was rooted with Phytopythiumvexansand Pythium undulatumas as the ourgroup. Likelihood of the best scoring ML tree was -114471.902046. Estimated base frequencies were as follows: A = 0.216570, C = 0.275568, G = 0.312230, T = 0.195632; substitution rates AC = 0.414835, AG = 1.176570, AT = 0.600142, CG = 0.970565, CT = 5.227735, GT = 1.000000.

No Comments