19 Sep Tilletia
Tilletia Tul. & C. Tul., Annls Sci. Nat., Bot., sér. 3 7: 112 (1847)
Background
Tulasne and Tulasne (1847) named Tilletia (Tilletiaceae, Exobasidiales) after Matthieu du Tillet (1714–1791), who first determined the pathogenicity of T. caries on wheat in France (Vánky and Shivas 2008). Tillet showed that washed seed reduced the spread of smut, although he was unaware the disease was caused by a fungus (Carefoot and Sprott 1967).
Species of Tilletia cause smut in the inflorescences and leaves of grasses (Poaceae). They are either localized in individual ovaries or systemic in the inflorescence. The species of Tilletia on cultivated grasses can cause economic losses and have been intensively studied. For example, several species, including T. caries, replace the grains of wheat with masses of spores that produce trimethylamine, which has an odour of rotten fish. Consequently, ginger-bread was invented as a solution to mask the smell of smutted grain (Carefoot and Sprott 1967). Tilletia indica is a billion-dollar threat to the wheat industries in Australia and the USA (Murray and Brennan 1998; Rossman 2009). The misidentification of T. indica in grain from both of these countries has been discussed (Castlebury and Carris 1999; Pascoe et al. 2005).
Castlebury et al. (2005) determined that the association of spore morphology, germination patterns and relationships with hosts were unclear in some clades of Tilletia. Systemic species of Tilletia germinate to form basidiospores that conjugate while attached to the basidium, and form dikaryotic hyphae that infect host seedlings. Localized species of Tilletia form basidiospores that do not conjugate on germination from the basidium (Castlebury et al. 2005). The systemic species usually have reticulate spores, whereas the localized species have verrucose spores (Castlebury et al. 2005).
Classification – Exobasidiomycetes, Exobasidiomycetidae, Tilletiales, Tilletiaceae
Type species – Tilletia caries (DC.) Tul. & C. Tul., Annls Sci. Nat., Bot., sér. 3 7: 113 (1847)
Distribution – Worldwide
Disease Symptoms – Sori mostly replace ovaries of infected grasses with a mass of powdery black or brown spores. The ovaries are often hypertrophied and the infection can be systemic or localised. Sori are sometimes produced on the leaves and culms of infected plants.
Hosts – Poaceae
Morphological based identification and diversity
Vánky (2011) listed 178 taxa in his world monograph of smut fungi. Since then only three further species, T. geeringii, T. mactaggartii and T. marjaniae, have been described, all from Australia on species of Eriachne (Li et al. 2014). Castlebury et al. (2005) found that Conidiosporomyces, Ingoldiomyces and Neovossia, collectively represented by only five species, were congeneric with Tilletia. Chandra and Huff (2008) established the monotypic Salmacisia, which was sister to Tilletia. Salmacisia buchloeana grouped with Tilletia, sister to T. dactyloctenii, in the present analysis. Vánky (2011) used host taxonomy as the most important character to identify species of Tilletia. The size and ornamentation of spores is used to further identify species on the same host genera (see http://collections.daff.qld.gov.au/web/key/smutfungi/ Shivas et al. 2014).
Molecular based identification and diversity
Castlebury et al. (2005) used the large subunit (LSU) region of ribosomal DNA (rDNA) to first study species of Tilletia with a molecular approach. Single species descriptions have since been based on the internal transcribed spacer (ITS) and LSU regions (Shivas et al. 2009; McTaggart and Shivas 2009; Li et al. 2014). The present study builds on work by previous authors and provides molecular barcodes for the ITS and LSU regions sequenced from type specimens in the private collection of Kálmán Vánky (Herbarium Ustliaginales Vánky), which is held at the Queensland Plant Pathology Herbarium (BRIP).
There are five publicly available genomes in GenBank for species of Tilletia, all of which are agriculturally important taxa. These are T. controversa, T. caries, T. walkeri (Nguyen et al. unpublished), T. horrida (Wang et al. 2015) and T. indica (Sharma et al. 2016). Their genomes range in size from 20–37 Mb with an average size of ~28 Mb. An 18 Mb genome for Tilletia buchloeana (as Salmacisia buchloeana) was recently released (Huff et al. 2017). An ongoing challenge for genomic studies of Tilletia is that these taxa are difficult to grow in culture, which limits the amount of DNA available for genomic analysis.
This study reconstructs the phylogeny of Tilletia based on analyses of combined ITS and LSU sequence data. The molecular barcodes of rDNA gene regions provided from type specimens in the present study will aid identification of known taxa. However, for further resolution within Tilletia, additional markers will be required. There was no phylogenetic support for the larger clades in the current analyses.
Recommended genetic marker (genus level) – LSU
Recommended genetic marker (species level) – ITS
Accepted number of species: There are 336 species epithets in Index Fungorum (2018) under this genus, of which 181 are in use.
References: Castlebury et al. 2005, Shivas et al. 2009, 2014, McTaggart and Shivas 2009, Vánky 2011, Li et al. 2014 (morphology and phylogeny); Wang et al. 2015, Sharma et al. 2016, Huff et al. 2017 (genome).
Table Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Taxon | Voucher number | ITS | LSU |
| Erratomyces patelii | HUV 18697 | DQ663692 | AF009855 |
| Oberwinkleria anulata | HUV 16003* | DQ875369 | NA |
| Salmacisia buchloëana | WSP 71313 | EF204936 | DQ659922 |
| Tilletia aegopogonis | WSP 67743 | AY818967 | NA |
| T. anthoxanthi | HUV 18739* | MH231773 | MH231773 |
| T. asperifolia | LMC 90 | NA | AY818968 |
| T. australiensis | BRIP 51874 | MH231774 | MH231774 |
| T. ayresii | HUV 19314 / BRIP 49130 | AY819017 | MH231775 |
| T. barclayana | Strain-832 | AF310168 | NA |
| T. barclayana | S-104 | AF399894 | NA |
| T. barclayana | WSP 68658 | NA | AY818970 |
| T. barclayana | WSP 68466 | NA | AY818971 |
| T. bornmuelleri | S 054 | AF398452 | NA |
| T. boutelouae | WSP 68661 | NA | AY818973 |
| T. brachypodii-mexicani | HUV 16007* | MH231776 | MH231776 |
| T. bromi | BRIP 49095 | MH231777 | MH231777 |
| T. capeyorkensis | BRIP 27011 | MH231778 | MH231778 |
| T. caries | LMC 97-136 | AF398438 | AY819007 |
| T. cerebrina | LMC 125 | NA | AY818994 |
| T. challinoriae | BRIP 52502* | NR119757 | NA |
| T. chionachnes | BRIP 26898* | MH231779 | MH231779 |
| T. controversa | V 764 | AF398440 | AY818995 |
| T. dactyloctenii | HUV 8887* | MH231780 | MH231780 |
| T. ehrhartae | BRIP 28392 | MH231781 | MH231781 |
| T. elymi | S 064 | AF398454 | NA |
| T. eragrostiellae | HUV 15805* | MH231782 | NA |
| T. eremopoae | HUV 19420* | MH231783 | MH231783 |
| T. filisora | BRIP 47729 | MH231784 | MH231784 |
| T. fusca | LMC 214 | AF398455 | AY818996 |
| T. geeringii | BRIP 51851* | KF055226 | NA |
| T. gigacellularis | HUV 20555* | MH231785 | MH231785 |
| T. goloskokovii | LMC 315 | NA | AY818999 |
| T. holci | V 765 | AF398459 | AY819008 |
| T. horrida | NA | AF398435 | NA |
| T. horrida | LMC 339 | NA | AY818974 |
| T. horrida | LMC 358 | NA | AY818975 |
| T. horrid | T54899 | MH231786 | NA |
| T. hyalospora | HUV 16038 | AF133576 | AF399891 |
| T. imbecillus | BRIP 7831 | MH231787 | MH231787 |
| T. indica | BPI 863665 | AF398434 | AY818977 |
| T. iowensis | BPI 863664 | NA | AY818988 |
| T. ischaemi | HUV17453* | MH231788 | MH231788 |
| T. ixophori | WSP 71170 | NA | AY819010 |
| T. kimberleyensis | BRIP 51857 | MH231789 | MH231789 |
| T. lachnagrostidis | BRIP 47300 | MH231790 | NA |
| T. laevis | V 766 | AF398444 | AY819005 |
| T. lageniformis | BRIP 47749* | MH231791 | MH231791 |
| T. laguri | HUV 16352* | MH231792 | NA |
| T. lineate | BRIP 26844* | MH231793 | MH231793 |
| T. lolii | S 119 | AF398460 | NA |
| T. maclaganii | Tm001NY09 | JF745116 | NA |
| T. mactaggartii | BRIP 51853* | KF055227 | KF055228 |
| T. majuscule | BRIP 51841* | NA | MH231794 |
| T. marjaniae | BRIP 49721* | KF055224 | KF055225 |
| T. menieri | WSP 69115 | AF398456 | AY819002 |
| T. micrairae | BRIP 52433* | FJ862995 | NA |
| T. moliniae | TUB 018922 | EU659134 | EU661605 |
| T. narayanaraoana | BRIP 47957 | GQ497894 | NA |
| T. nigrifaciens | BRIP 43865 | MH231796 | MH231796 |
| T. obscura-reticulata | WSP 68357 | NA | AY819011 |
| T. olida | BRIP 44536 | MH231797 | MH231797 |
| T. opaca | BRIP 27896 | MH231798 | MH231798 |
| T. panici-humilis | HUV 205832* | MH231799 | NA |
| T. polypogonis | V 931 | NA | AY819015 |
| T. pseudochaetochloae | BRIP 46730 | MH231800 | MH231800 |
| T. pseudoraphidis | BRIP 51873* | MH231801 | MH231801 |
| T. pulcherrima | WSP 71501 | EU915293 | NA |
| T. rostrariae | HUV 14898* | MH231802 | MH231802 |
| T. rugispora | HUV 19147 / BRIP 47127 | MH231803 | AY818983 |
| T. savilei | V 859 | AF399885 | AY819018 |
| T. sehimicola | BRIP 51847* | MH231804 | MH231804 |
| T. setariae | V 934 | NA | AY819014 |
| T. setariae-parvifolia | BRIP 47735* | MH231805 | NA |
| T. setariae-pumilae | HUV 21399* | MH231806 | NA |
| T. shivasii | BRIP 52525 | MH231807 | MH231807 |
| T. sporoboli | HUV 1880* | MH231808 | NA |
| T. sterilis | LMC 363 | NA | AY819003 |
| T. sumatiae | HUV 17529 / V933 | MH231809 | AY818987 |
| T. thailandica | BRIP 48134 | NA | MH231810 |
| T. trabutii | BRIP 46328 | MH231811 | MH231811 |
| T. trachypogonis | HUV 19626* | MH231812 | MH231812 |
| T. triticoides | S 102 | AF398446 | NA |
| T. verruculosa | WSP 70430 | NA | AY818984 |
| T. viennotii | BRIP 47077 | MH231813 | MH231813 |
| T. vitatta | BRIP 54207 | MH231814 | MH231814 |
| T. walker | BPI 746091 | AF399887 | AY818978 |
| T. whiteochloae | BRIP 51838 | MH231815 | MH231815 |
| T. xerochloae | BRIP 54437 | MH231816 | MH231816 |
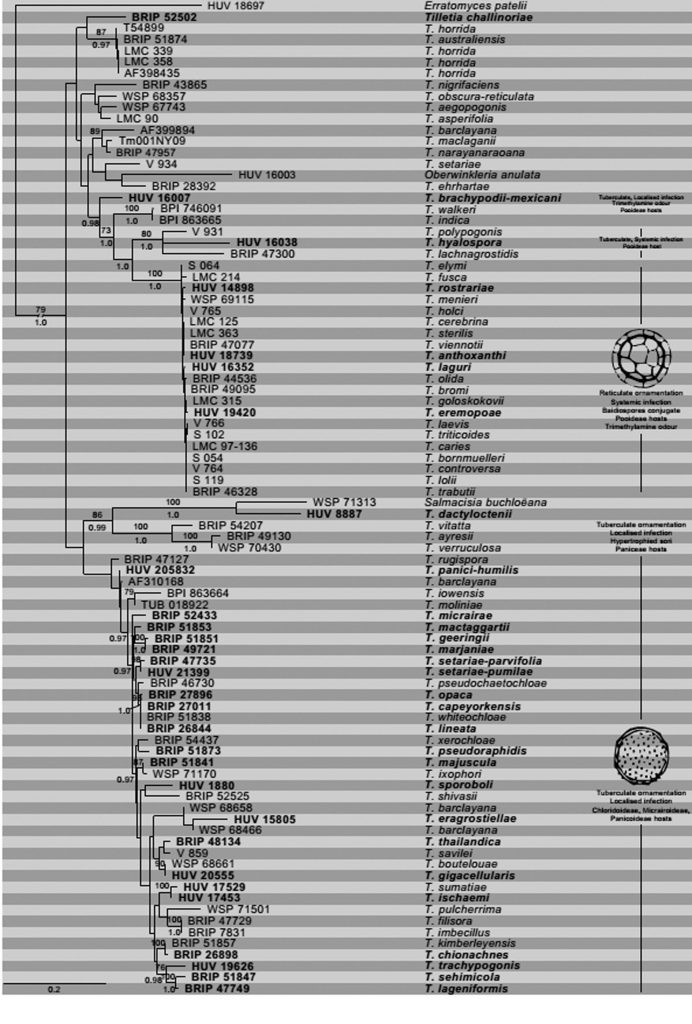 Fig. Phylogram of 89 species of Tilletia obtained from a maximum likelihood search (command –f a) of concatenated ITS and LSU gene regions in RAxML v. 8.2 (Stamatakis 2014). Bootstrap values (≥70%) from 1000 maximum likelihood replicates above nodes and posterior probability values (≥0.95) summarized from 30,000 converged trees in a Bayesian search below nodes. Taxa sequenced from a type specimen in bold font. The tree was rooted to Erratomyces patellii.
Fig. Phylogram of 89 species of Tilletia obtained from a maximum likelihood search (command –f a) of concatenated ITS and LSU gene regions in RAxML v. 8.2 (Stamatakis 2014). Bootstrap values (≥70%) from 1000 maximum likelihood replicates above nodes and posterior probability values (≥0.95) summarized from 30,000 converged trees in a Bayesian search below nodes. Taxa sequenced from a type specimen in bold font. The tree was rooted to Erratomyces patellii.

No Comments