15 Oct Pestalotiopsis
Pestalotiopsis Steyaert, Bull. Jard. bot. État Brux. 19: 300 (1949)
Pestalotiopsis is an appendage-bearing, 5-celled conidia (asexual coelomycetes) in the family Sporocadaceae (Maharachchikumbura et al. 2014a, b; Jayawardena et al. 2016). The genus was introduced by Steyaert (1949). Pestalotiopsis species are widely distributed throughout tropical and temperate regions (Guba 1961; Maharachchikumbura et al. 2012, 2014a). Pestalotiopsis species have been isolated from dead leaves, bark, twigs, soil, polluted stream water, wood, paper, fabrics, and wool (Guba 1961; Maharachchikumbura et al. 2012, 2014a). Some species have been associated with human and animal infections, and others (e.g. P. guepinii and P. microspora) have also been isolated from extreme environments (Maharachchikumbura et al. 2014b).
Classification – Sordariomycetes, Xylariomycetidae, Amphisphaeriales, Sporocadaceae
Type species – Pestalotiopsis guepinii (Desm.) Steyaert [as ‘guepini’], Bull. Jard. bot. État Brux. 19(3): 312 (1949)
Distribution – Worldwide
Disease symptoms – Species of Pestalotiopsis cause a variety of diseases in plants including canker lesions, shoot dieback, leaf spots, needle blight, tip blight, grey blight, scabby canker, severe chlorosis, fruit rots and various post-harvest diseases (Maharachchikumbura et al. 2013a, b, 2014a, b). These pathogens reduce production and cause economic loss in apple, blueberry, coconut, chestnut, ginger, grapevine, guava, hazelnut, lychee, mango, orchid, peach, rambutan, tea and wax apple due to diseases (Maharachchikumbura et al. 2013a, b, 2014a, b). Grapevine trunk diseases are the most destructive diseases of grapevines that impact the economic production and longevity of vineyards and even leading to partial or total death of individual plants. Therefore, the initial identification of the causal agent is essential for early control of these diseases (Jayawardene et al. 2015; Maharachchikumbura et al. 2017). Pestalotoid fungi have been reported as pathogens on a variety of grapevine cultivars, causing diseases including grapevine dieback, fruit rot, postharvest disease and severe defoliation and they infect all plant parts including leaves, canes, wood, berries and flowers (Jayawardene et al. 2015; Maharachchikumbura et al. 2017). Pestalotiopsis menezesiana (Bres. & Torr.) Bissett. and P. uvicola (Spegazzini) Bissett, are the most common species recorded from grapevine around the world and especially P. biciliata are associated with trunk grapevine disease (Jayawardene et al. 2015; Maharachchikumbura et al. 2017).
Hosts– Broad range of hosts including members of Altingiaceae, Arecaceae, Bromeliaceae, Euphorbiaceae, Myrtaceae, Poaceae, Proteaceae, Rosaceae, Rutaceae, Theaceae and Vitaceae.
Morphological based identification and diversity
There are around 250 species, most of which were named according to their host associations (Maharachchikumbura et al. 2014a, b). However, Pestalotiopsis species are not hosted specific and are found on a wide range of plants and substrates (Jeewon et al. 2003; Lee et al. 2006; Maharachchikumbura et al. 2014a, b). They exhibit considerable diversity in phenotype, and group together based on similarities in conidial morphology (Jeewon et al. 2003; Maharachchikumbura et al. 2012, 2013a, b, 2014a, b). Considering morphology, conidial length, width, median cell length, the colour of median cells and length of the apical appendages appear to be stable characters within Pestalotiopsis (Jeewon et al. 2003; Maharachchikumbura et al. 2014b).
Pestalotiopsis guepinii was considered to be the type species of the genus described from stems and leaves of Camellia japonica collected in France, and is characterised by 5-celled conidia with three concolourous median cells, hyaline terminal cells and simple or unbranched appendages arising from the apex of the apical cell (Steyaert 1949; Maharachchikumbura et al. 2014b). Nag Raj (1985) regarded P. maculans as the type species of Pestalotiopsis with P. guepinii as a synonym. Jeewon et al. (2003) also accepted P. maculans clusters with species having concolourous median cells based on phylogenetic analysis of ITS sequence data and that P. karstenii might be a synonym of P. maculans (Maharachchikumbura et al. 2014b).
Most Pesalotiopsis species lack sexual morphs. The sexual morph of Pestalotiopsis was treated as Pestalosphaeria Barr, with the type species Pestalosphaeria concentrica collected from grey-brown spots on living leaves of Rhododendron maximum in North Carolina, USA (Maharachchikumbura et al. 2014b). Pestalosphaeria concentrica is characterised by immersed, subglobose ascomata and unitunicate, cylindrical asci with a J+ apical ring; ascospores uniseriate in the ascus, ellipsoid, pale dull brown and 2-septate (Maharachchikumbura et al. 2014b).
Pestalotiopsis species have the ability to switch life-modes as endophytes, pathogens and saprobes (Hu et al. 2007; Maharachchikumbura et al. 2012). Therefore, many endophytic and plant pathogenic Pestalotiopsis species persist as saprobes and have been isolated from dead leaves, bark and twigs (Maharachchikumbura et al. 2012, 2013a, b, 2014b).
Pestalotiopsis species that were isolated as endophyte are important in the production of novel compounds with medicinal, agricultural and industrial applications (Maharachchikumbura et al. 2014b; Xu et al. 2010, 2014). Pestalotiopsis species are a rich source for bioprospecting compared to other fungal genera, and more than 100 compounds have been isolated from Pestalotiopsis (Maharachchikumbura et al. 2014b; Xu et al. 2010, 2014).
Molecular based identification and diversity
Maharachchikumbura et al. (2012) tested with 10 gene regions to resolve species boundaries in Pestalotiopsis (actin, calmodulin, glutamine synthase, glyceraldehyde-3-phosphate dehydrogenase, ITS, LSU, 18S nrDNA, RNA polymerase II, tef1 and TUB2). Maharachchikumbura et al. (2014b) used phylogenetic analysis of combined ITS, TUB2 and tef1 genes to successfully resolve Pestalotiopsis species.
Recommended genetic markers (genus level) – LSU (as outlined in Maharachchikumbura et al. 2012)
Recommended genetic markers (species level) – ITS, TUB2 and tef1 (as outlined in Maharachchikumbura et al. 2012)
Accepted number of species: There are 360 epithets in Index Fungorum in this genus, however, 75 species with DNA sequence data are accepted.
References: Maharachchukumbura 2013, 2014b, 2016b (morphology, phylogeny)
Table Details of Pestalotiopsis the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher No | ITS | TUB2 | tef1 |
| Pestalotiopsis adusta | ICMP 6088* | JX399006 | JX399037 | JX399070 |
| P. aggestorum | LC6301* | KX895015 | KX895348 | KX895234 |
| P. anacardiacearum | IFRDCC 2397* | NR120255.1 | KC247155 | KC247156 |
| P. arceuthobii | CBS 434.65* | NR147561 | KM199427 | KM199516 |
| P. arengae | CBS 331.92* | NR147560 | KM199426 | KM199515 |
| P. australasiae | CBS 114126* | NR147546 | KM199409 | KM199499 |
| P. australis | CBS 114193* | KM199332 | KM199383 | KM199475 |
| P. biciliata | CBS 124463* | KM199308 | KM199399 | KM199505 |
| P. brachiata | LC2988* | KX894933 | KX895265 | KX895150 |
| P. brassicae | CBS170.26* | NR147562 | – | KM199558 |
| P. camelliae | MFLUCC 12-0277* | NR120188 | JX399041 | JX399074 |
| P. chamaeropis | CBS 186.71* | KM199326 | KM199391 | KM199473 |
| P. clavata | MFLUCC 12-0268* | JX398990 | JX399025 | JX399056 |
| P. colombiensis | CBS 118553* | NR147551 | KM199421 | KM199488 |
| P. digitalis | ICMP 5434* | KP781879 | KP781883 | – |
| P. dilucida | LC3232* | KX894961 | KX895293 | KX895178 |
| P. diploclisiae | CBS 115587* | NR147552 | KM199419 | KM199486 |
| P. diversiseta | MFLUCC 12-0287* | NR120187 | JX399040 | JX399073 |
| P. dracontomelon | MFUCC 10-0149* | KP781877 | – | KP781880 |
| P. ericacearum | IFRDCC 2439* | KC537807 | KC537821 | KC537814 |
| P. formosana | NTUCC 17-009* | MH809381 | MH809385 | MH809389 |
| P. furcata | MFLUCC 12-0054* | JQ683724 | JQ683708 | JQ683740 |
| P. gaultheria | IFRD 411-014* | KC537805 | KC537819 | KC537812 |
| P. gibbosa | NOF3175* | LC311589 | LC311590 | LC311591 |
| P. grevilleae | CBS 114127* | NR147548 | KM199407 | KM199504 |
| P. hawaiiensis | CBS 114491* | NR147559 | KM199428 | KM199514 |
| P. hollandica | CBS 265.33* | KM199328 | KM199388 | KM199481 |
| P. humus | CBS 336.97* | KM199317 | KM199420 | KM199484 |
| P. inflexa | MFLUCC 12-0270* | JX399008 | JX399039 | JX399072 |
| P. intermedia | MFLUCC 12-0259* | JX398993 | JX399028 | JX399059 |
| P. italiana | MFLUCC 12-0657* | KP781878 | KP781882 | KP781881 |
| P. jester | CBS 109350* | KM199380 | KM199468 | KM199554 |
| P. jiangxiensis | LC4399* | KX895009 | KX895341 | KX895227 |
| P. jinchanghensis | LC6636* | KX895028 | KX895361 | KX895247 |
| P. kenyana | CBS 442.67* | KM199302 | KM199395 | KM199502 |
| P. knightiae | CBS 114138* | KM199310 | KM199408 | KM199497 |
| P. licualacola | HGUP 4057* | KC492509 | KC481683 | KC481684 |
| P. linearis | MFLUCC 12-0271* | JX398992 | JX399027 | JX399058 |
| P. longiappendiculata | LC3013* | KX894939 | KX895271 | KX895156 |
| P. lushanensis | LC4344* | KX895005 | KX895337 | KX895223 |
| P. macadamiae | BRIP 63738b* | KX186588 | KX186680 | KX186621 |
| P. malayana | CBS 102220* | NR147550 | KM199411 | KM199482 |
| P. monochaeta | CBS 144.97* | KM199327 | KM199386 | KM199479 |
| P. montellica | MFLUCC 12-0279* | JX399012 | JX399043 | JX399076 |
| P. neolitseae | NTUCC 17-011* | MH809383 | MH809387 | MH809391 |
| P. novae-hollandiae | CBS 130973* | NR147557 | KM199425 | KM199511 |
| P. olivaceae | PSHI2002* | AY687883 | DQ333580 | – |
| P. oryzae | CBS 353.69* | KM199299 | KM199398 | KM199496 |
| P. pallidotheae | MAFF 240993* | NR111022 | LC311584 | LC311585 |
| P. papuana | CBS 887.96* | KM199318 | KM199415 | KM199492 |
| P. parva | CBS 265.37* | KM199312 | KM199404 | KM199508 |
| P. photinicola | GZCC 16-0028* | KY092404 | KY047663 | KY047662 |
| P. portugalica | CBS 393.48* | KM199335 | KM199422 | KM199510 |
| P. rhizophorae | MFLUCC 17-0416* | MK764283 | MK764349 | MK764327 |
| P. rhododendri | IFRDCC 2399* | NR120265 | KC537818 | KC537811 |
| P. rhodomyrtus | HGUP 4230* | KF412648 | KF412642 | KF412645 |
| P. rosea | MFLUCC 12-0258* | JX399005 | JX399036 | JX399069 |
| P. scoparia | CBS 176.25* | KM199330 | KM199393 | KM199478 |
| P. shorea | MFLUCC 12-0314* | KJ503811 | KJ503814 | KJ503817 |
| P. spathulata | CBS 356.86* | NR147558 | KM199423 | KM199513 |
| P. telopeae | CBS 114161* | NR147545 | KM199403 | KM199500 |
| P. thailandica | MFLUCC 17-1616* | MK764285 | MK764351 | MK764329 |
| P. trachicarpicola | IFRDCC 2440* | NR120109 | JQ845945 | JQ845946 |
| P. unicolor | MFLUCC 12-0276* | JX398999 | JX399030 | – |
| P. verruculosa | MFLUCC 12-0274* | NR120185 | – | JX399061 |
| P. yanglingensis | LC4553* | KX895012 | KX895345 | KX895231 |
| P. yunnanensis | HMAS 96359* | AY373375 | – |
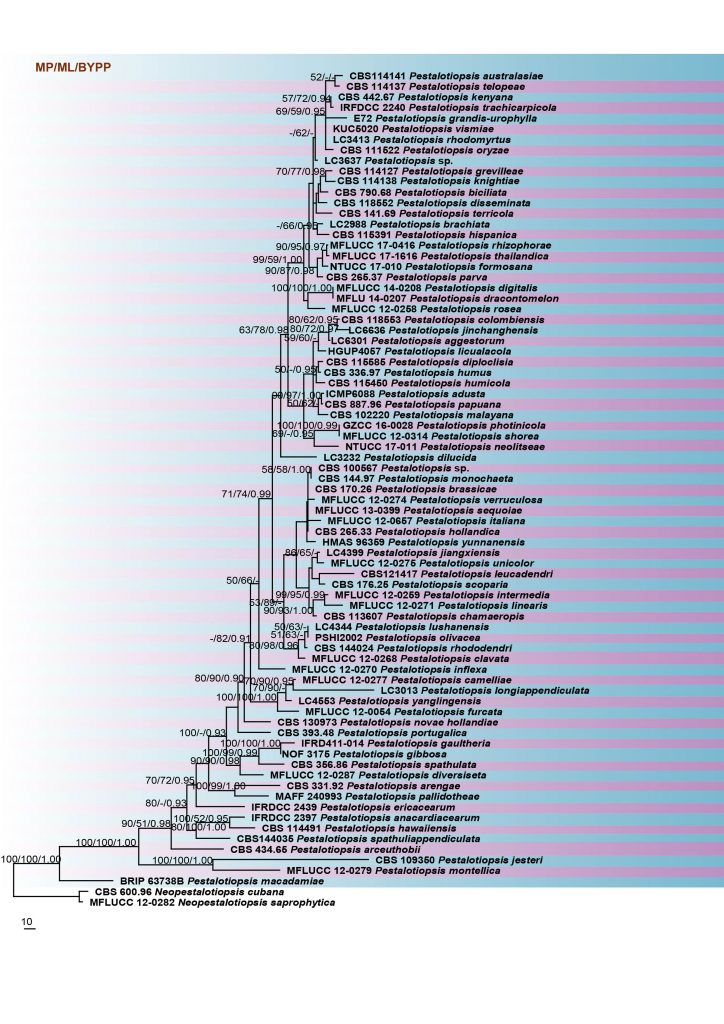
Fig. Phylogram generated from RAxML analysis based on combined ITS, TUB2 and tef1 sequences of all the accepted species of Pestalotiopsis. Related sequences were obtained from GenBank. Seventy-nine taxa are included in the analyses, which comprise 1581 characters including gaps. The tree was rooted in Neopestalotiopsis cubana (CBS 600.96) and N. saprophytica (MFLUCC 12-0282). Tree topology of the ML analysis was similar to the BYPP and MP. The best scoring RAxML tree with a final likelihood value of -12269.881063 is presented. The matrix had 763 distinct alignment patterns, with 15.79% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.234129, C = 0.293516, G = 0.211518, T = 0.260837; substitution rates AC = 1.189917, AG = 3.402399, AT = 1.153875, CG = 1.001451, CT = 4.301074, GT = 1.000000; gamma distribution shape parameter α = 0.283001. The maximum parsimonious dataset consisted of 948 constant, 450 parsimony-informative and 183 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of ten equally most parsimonious trees with a length of 1962 steps (CI = 0.498, RI = 0.697, RC = 0.347, HI = 0.502) in the first tree. RAxML and maximum parsimony bootstrap support value ≥50% and BYPP ≥0.90 values are shown near the nodes. The scale bar indicates 10 changes per site. The ex-type strains are in bold.

No Comments