26 Oct Heterobasidion
Heterobasidion Bref., Unters. Gesammtgeb.Mykol. (Liepzig) 8: 154 (1888)
Background
Heterobasidion was introduced by Brefeld (1888) and is typified by H. annosum (≡ Polyporus annosus). Certain Heterobasidion species are important forest pathogens of the Northern Hemisphere, causing root and butt rot, mainly in conifers (Woodward et al. 1998). In coniferous plantations, Heterobasidion is one of the most widespread of wood decay agents, especially when the host is under intensive management. Heterobasidion greatly reduces site productivity and the amount of harvestable timber; estimated financial losses caused by Heterobasidion species in Europe were around 800 million euro per year (Korhonen et al. 1998; Garbelotto 2004; Asiegbu et al. 2005). On the other hand, these taxa have a relatively moderate pathogenic role in natural forest ecosystems. They affect stand species composition, density and structure, and they contribute to forest succession, nutrient recycling and even regeneration (Goheen and Otrosina 1998; Garbelotto 2004; Dai et al. 2006).
Classification – Basidiomycota, Agaricomycotina, Agaricomycetes, Incertaesedis, Russulales, Bondarzewiaceae
Type species – Heterobasidion annosum (Fr.) Bref., Unters. Gesammtgeb. Mykol. (Liepzig) 8: 154 (1888)
Distribution – North America, Europe, Asia, Australia and Oceania
Disease symptoms – There are two Heterobasidion species complexes –H. insulare sensu lato and H. annosum sensu lato – they cause the same symptoms. The H. annosum species complex is one of the major root-rot pathogenic genera of the northern temperate hemisphere (Garbelotto and Gonthier 2013; Kärhä et al. 2018). After the primary infection through stump tops, or stem and root wounds, the taxa can vegetatively infect uninjured trees (secondary infection) by the growth of the mycelium through root contacts (Rishbeth 1950, 1951a,b; Asiegbu et al. 2005; Garbelotto and Gonthier 2013). Heterobasidion could be considered both necrotrophs and saprotrophs; though some species in the H. insulare species complex (e.g. H. austral, H. araucariae) are mainly saprotrophs (Niemelä and Korhonen 1998; Dai and Korhonen 2009; Chen et al. 2014). In contrast to Europe, the pathogenicity of H. annosum sensu lato in China and Japan is uncertain; the complex seems to occur mostly on dead trees, and no symptoms of tree decline are usually visible near infected trees. These observations could be due to different, less intensive forest management strategies in the East-Asian regions, or lack of data on the butt rot symptoms (Dai et al. 2006; Tokuda et al. 2007).
The infection causes white pocket rot and heart rot in the roots and the butt of living trees (Korhonen and Stenlid 1998; Asiegbu et al. 2005). Resin, containing mycelium, may also exude from the infected roots, or the bark-scales (Rishbeth 1950). In invaded roots and the basal portions of the trunk, H. annosum sensu lato taxa colonize different plant tissues depending on the host. Heart rot is mainly caused in trees more susceptible to the colonization of the heartwood, e.g. Picea abies. In the case of Pinus, cambium and sapwood are the most severely colonized, while the sapwood of Calocedrus or Sequoiadendron trees is the most colonized (Garbelotto 2004; Asiegbu et al. 2005; Garbelotto and Gonthier 2013).
After establishment, the basidiomata of H. annosum sensu lato appear. The localization of the sporocarps is governed by the species, environmental conditions and infection strategy. Some species prefer the root collar for fruiting (H. annosum, H. irregular). Some also produce sporocarps in decay pockets in stumps and fallen trees (H. parviporum, H. abietinum and H. occidentale), or under the intact surface of stumps (H. irregulare, H. occidentale). The sporocarps are sometimes located on the higher parts of the trunk. When moisture is limited, the fungi fruit inside stumps; if the climate is moist and humid, the basidiomata can be found near the ground in the duff at the base of diseased trees. If during primary infection the stump surface is infected, the basidiomata form under an intact top layer. During active pathogenesis, if the standing trees are infected the sporocarps could be found within decay columns in the sapwood (Rishbeth 1950; Otrosina and Garbelotto 2010).
The infection kills the functioning sapwood, cambium and heart wood in the roots and at the basal portions of the trunk, resulting in white rot, reduced growth rate, crown dieback (Omdal et al. 2004), and eventually mortality and windthrow of infected trees (Rishbeth 1950; Olivaet al. 2008; Garbelotto and Gonthier 2013).
Hosts –The host range of Heterobasidion is extremely wide. The genus has been reported from approximately 200 host species (Korhonen and Stenlid 1998). Taxa mostly occur on gymnosperms, such as Abies, Agathis, Araucaria, Calocedrus, Juniperus, Keteleeria, Larix, Picea, Pinus, Podocarpus, Pseudolarix, Pseudotsuga, Sequoia, Sequoiadendron, Thuja and Tsuga (Buchanan 1988; Corner 1989; Dai and Korhonen 2009; Otrosina and Garbelotto 2010; Garbelotto and Gonthier 2013; Garbelotto et al. 2017). Occasionally, certain Heterobasidion species grow on broad-leaved trees of various angiosperm genera (Garbelotto and Gonthier 2013; Ryvarden and Melo 2014).
Morphological based identification and diversity
There are 33 Heterobasidion epithets listed in Index Fungorum (2020). Of these, eight are related to other polypore genera, based on type studies and morphological observations (Ryvarden 1972, 1985; Buchanan and Ryvarden 1988; Dai and Niemelä1995; Hattori 2003). Besides, the taxonomic status of three further species described from Asia is unclear: viz. H. arbitrarium, H. perplexum and H. insulare (Corner 1989; Ryvarden 1989; Stalpers 1996; Hattori 2001; Dai et al. 2002; Tokuda et al. 2009). Given that no sequence data (H. arbitrarium, H. perplexum) or authentic sequences (H. insulare sensu stricto) are available for the molecular resolution, further studies are needed to clarify their status.
Formerly, Heterobasidion was considered as a group consisting of only the generic type, H. annosum and H. araucariae and H. insulare (Buchanan 1988; Chase 1989). However, mating studies on Eurasian and North American Heterobasidion collections revealed several intersterile groups, which later became the basis for designating separate taxonomic species within the H. annosum and H. insulare species complexes. Mating experiments revealed three intersterile groups of H. annosum sensu lato in Europe (Korhonen 1978b, Capretti et al. 1990) and two in North America (Otrosina et al. 1993). All intersterile groups have been recognised in the H. annosum species complex are now formally described as separate taxonomic species. European groups were described as H. abietinum, H. parviporum and H. annosum sensu stricto (Niemelä and Korhonen 1998), whereas North American groups were named H. irregulare and H. occidentale (Otrosina and Garbelotto 2010).
The mating study by Dai et al. (2002) on Asian “H. insulare” collections revealed three intersterile groups in China, which were subsequently described as Heterobasidion linzhiense (Dai et al. 2007), H. orientale and H. ecrustosum (Tokuda et al. 2009). H. australe related to the H. insulare species complex was also described from China by Dai and Korhonen (2009). Chen et al. (2014) described two further Heterobasidion species (H. amyloideum and H. tibeticum) from the eastern Himalayas based on phylogenetic evidence. These species are morphologically closely related to the members of the H. insulare species complex, but differ in presence of cystidia and amyloid skeletal hyphae in the context. The recently described H. amyloideopsis was collected in the western Himalayas (Pakistan) and formed a monophyletic group with the H. insulare species complex, sister to H. amyloideum (Zhao et al. 2017).
The main morphological characters which are used for the identification are the resupinate to pileate basidiocarps, the dimitic hyphal system with mostly simple septate generative hyphae, and the asperulate basidiospores showing no reaction in Melzer’s reagent. Besides morphology, host preference, geographical distribution, and DNA sequence data have also been used for species identification (Otrosina and Garbelotto 2010; Chen et al. 2015a).
Fig. 1 Members of Heterobasidion annosum species complex. a basidiome on Scots pine, b basidiome on European silver fir, c–e basidiomes on European spruce, f hyphal structure in the trama, g hyphal structure in the context, h–j basidiospores. Scale bars: f–g = 10 µm, h–j = 5 µm
Molecular based identification and diversity
Heterobasidion has been intensely studied by molecular methods. Sequence data are available for the majority of taxa, and molecular studies were conducted to understand the evolution (Dalman et al. 2010), mating behaviour (Gonthier and Garbelotto 2011), and pathogenicity (Liu et al. 2018a) of Heterobasidion species.
Various marker types were used to resolve the phylogeny of the H. annosum species complex, such as isoenzyme (Karlsson and Stenlid 1991), AFLP (Gonthier and Garbelotto 2011) and SSR (Garbelotto et al. 2013) markers. Sequence analyses were carried out initially on nrITS and intergenic spacer regions (Kasuga and Mitchelson 1993; DeScenzo and Harrington 1994), housekeeping genes (Johanesson and Stenlid 2003), peroxidase (Maijala et al. 2003) and laccase genes (Asiegbu et al. 2004), with which it was possible to distinguish four lineages (three European and one North American) within the complex (Asiegbu et al. 2005). Later, allowing the differentiation of a larger number of taxa, further nuclear genes were applied, such as the calmodulin (cam), translation elongation factor 1-α (tef1), glyceraldehydes3-phosphate dehydrogenase (gapdh), heat shock protein (hsp), glutathione-S-transferase (gst1) and transcription factor (tf) genes (Johanesson and Stenlid 2003; Ota et al. 2006; Dalman et al. 2010), as well as two mitochondrial genes, the mitochondrial ATP synthase subunit 6 (ATP6) and mitochondrial rDNA region (Linzer et al. 2008). Dalman et al. (2010) came to the conclusion, that there are two monophyletic sister clades within the H. annosum species complex, representing the Eurasian and North American species.
The protein-coding largest subunit of RNA polymerase II (rpb1) and the second subunit of RNA polymerase II (rpb2) genes were used by Chen et al. (2014) and were suitable to differentiate Heterobasidion species in the H. insulare species complex. The variability of these markers was confirmed by Chen et al. (2015a) and Zhao et al. (2017) who, among other previously mentioned markers, both used the nuclear large ribosomal subunit (nrLSU) and the mitochondrial small subunit (mtSSU) sequences to their studies.
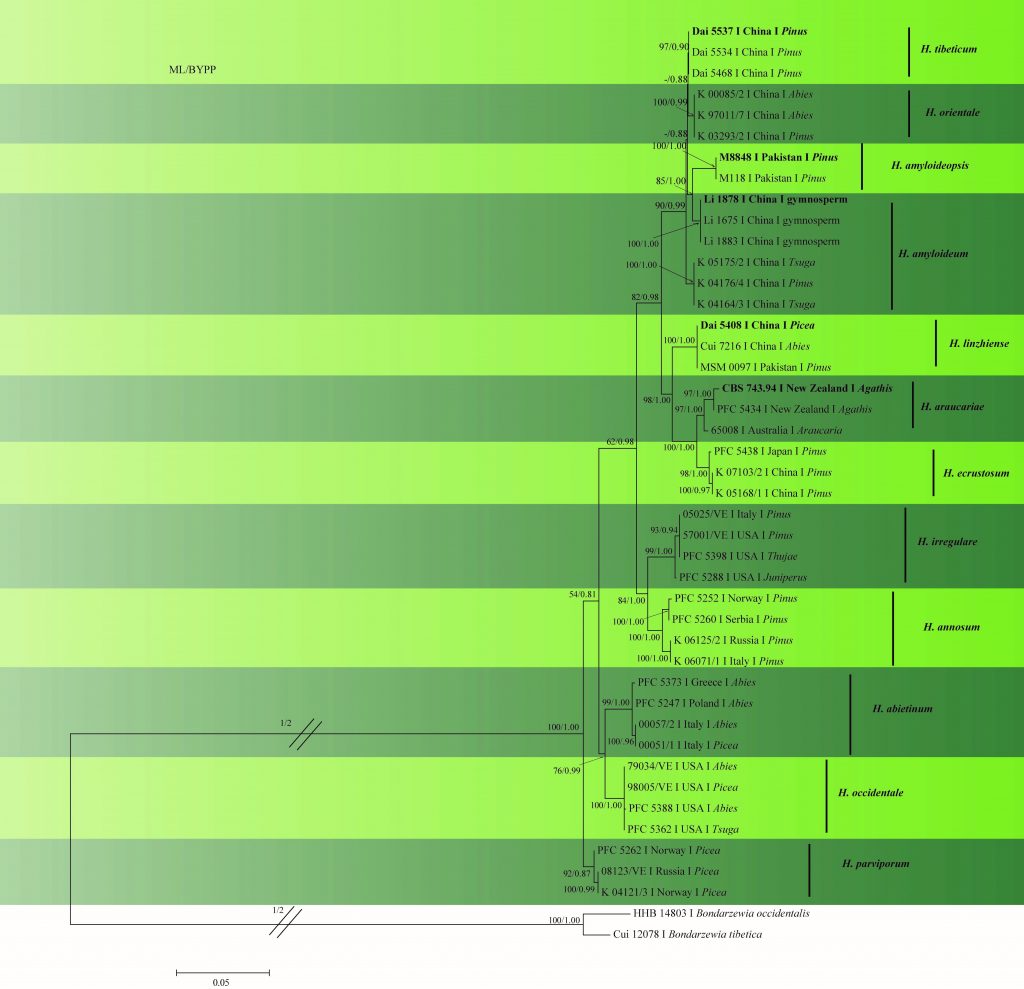
In this study, we provide a phylogenetic tree (Fig. 2) based on multi-locus phylogenetic analysis of ITS–gapdh–rpb1–rpb2–tef1 sequence data. Sequences of H. arbitrarium and H. perplexum could not be analyzed as they are unavailable in GenBank. Furthermore, no sequences are available for the type of H. insulare hence this species was not included in the analysis. The results provide a similar topology to those obtained by Chen et al. (2015) and Zhao et al. (2017).
Recommended genetic marker (genus level) – nLSU
Recommended genetic markers (species level) – rpb1, rpb2
Accepted number of species –There are 33 epithets in Index Fungorum (2020), however only 15 species are accepted (Table 1). Amongst these, no sequences are available for H. arbitrarum and H. insulare. Heterobasidion perplexum is not accepted in the genus, pending further studies.
References – Dai and Korhonen (2009) (new sp., China, morphology); Tokuda et al. (2009) (new species, East Asia); Dalman et al. (2010) (Evolution, H. annosum species complex, haplotype network); Otrosina and Garbelotto (2010) (new species, North America, biology); Garbelotto and Gonthier (2013) (biology, epidemiology, control); Chen et al. (2014) (new species, China, phylogeny); Chen et al. (2015a) (biogeography, divergence time estimation, phylogeny); Zhao et al. (2017) (new sp., Pakistan, phylogeny).
Table 1 DNA barcodes for accepted species of Heterobasidion. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold.
| Species | Strain | ITS | rpb1 | rpb2 | gapdh | tef1 |
| Heterobasidion abietinum | 00057/2 | KJ651453 | KJ651632 | KJ651725 | KJ651756 | – |
| 00051/1 | KJ651450 | KJ651629 | KJ651722 | KJ651753 | – | |
| PFC 5247 | KC492895 | – | – | KP863657 | KC571636 | |
| PFC 5373 | KC492956 | – | – | KP863664 | KC571687 | |
| H. amyloideopsis | MS 8848* | KT598384 | – | KT598388 | – | – |
| M 118 | KT598385 | – | KT598389 | – | – | |
| H. amyloideum | Li 1878* | KJ651455 | KF033157 | KF006538 | KJ651758 | – |
| Li 1675 | KJ651454 | KF033155 | KF006534 | KJ651757 | – | |
| Li 1883 | KJ651456 | KF033156 | KF006537 | KJ651759 | – | |
| H. annosum | K 06125/2 | KJ651459 | KF453492 | KF453498 | KJ651762 | – |
| K 06071/1 | KJ651458 | KF453491 | KF453497 | KJ651761 | – | |
| PFC 5252 | KC492906 | – | – | KP863659 | KC571646 | |
| PFC 5260 | KC492911 | – | – | KP863661 | KC571651 | |
| H. araucariae | 65008 | KJ651462 | KJ651636 | KJ651729 | KJ651766 | – |
| CBS 743.94* | MH862503 | – | – | – | – | |
| PFC 5434 | KX130098 | – | – | KX130104 | KX130101 | |
| H. australe | K 05175/2 | KJ651467 | KF033137 | KF006506 | KJ651771 | – |
| K 04167/4 | KJ651465 | KF033135 | KF006502 | KJ651769 | – | |
| K 04164/3 | KJ651464 | KF033134 | KF006500 | KJ651768 | – | |
| H. ecrustosum | K 05168/1 | KJ651468 | KF033142 | KF006513 | KJ651772 | – |
| K 07103/2 | KJ651471 | KF033145 | KF006517 | KJ651775 | – | |
| PFC 5438 | KX130099 | – | – | KX130105 | KX130102 | |
| H. irregulare | 57001/VE | KJ651473 | KJ651638 | KJ651731 | KJ651777 | – |
| 05025/VE | KJ651478 | KJ651643 | KJ651736 | KJ651782 | – | |
| PFC 5398 | KP863586 | – | – | KP863652 | KP863616 | |
| PFC 5288 | KP863575 | – | – | KP863641 | KP863606 | |
| H. linzhiense | Dai 5408* | KJ651484 | KF033154 | KF006533 | KJ651788 | – |
| Cui 7216 | KJ651480 | KF033148 | KF006524 | KJ651784 | – | |
| MSM 0097 | MH233930 | – | – | – | – | |
| H. occidentale | 79034/VE | KJ651485 | KJ651645 | KJ651738 | KJ651789 | – |
| 98005/VE | KJ651489 | KJ651649 | KJ651742 | KJ651793 | – | |
| PFC 5362 | KP863584 | – | – | KP863650 | KP863614 | |
| PFC 5388 | KP863585 | – | – | KP863651 | KP863615 | |
| H. orientale | K 97011/7 | KJ651490 | KF033141 | KF006512 | KJ651794 | – |
| K 03293/2 | KJ651493 | KF033140 | KF006510 | KJ651797 | – | |
| K 00085/2 | KJ651492 | KF033139 | KF006508 | KJ651796 | – | |
| H. parviporum | K 04121/3 | KJ583212 | KF453493 | KF453499 | KJ651800 | – |
| 08123/VE | KJ651500 | KF453495 | KF453501 | KJ651805 | – | |
| PFC 5262 | KC492957 | – | – | KP863662 | KC571688 | |
| H. tibeticum | Dai 5537* | KJ651507 | KF033153 | KF006531 | KJ651812 | – |
| Dai 5468 | KJ651505 | KF033151 | KF006527 | KJ651810 | – | |
| Dai 5534 | KJ651506 | KF033152 | KF006529 | KJ651811 | – |
Fig. 2 Phylogram generated from ML analysis based on combined ITS, rpb1, rpb2, gapdh and tef1 sequence data of Heterobasidion species. Related sequences were obtained from GenBank. Fourty-four strains are included in the analyses, which comprised 4314 characters including gaps. The tree was rooted with Bondarzewia occidentalis (HHB 14803) and B. tibetica (Cui 12078). Tree topology of the ML analysis was similar to to the Bayesian analysis. ML bootstrap values ˃50% and BYPP˃0.80 are shown respectively near the nodes.


No Comments