24 Oct Cercospora
Cercospora Fresen. ex Fuckel, Hedwigia 2(15): 133 (1863)
Background
Cercospora includes pathogens, saprobes and endophytes. Species are widely distributed, occurring on numerous flowering and ornamental plants, ferns, other fungi (as parasites), gymnosperms, grasses and other monocotyledons such as lilies, magnoliids and palms, mostly causing leaf spots. The well-known asexual morph, which is hyphomycetous, are among the largest groups of plant pathogenic fungi causing leaf spots, leading to diseases on many economically important crops (Agrios 2005; To-Anun et al. 2011; Groenewald et al. 2013; Guatimosim et al 2016; Park et al. 2017). Comparatively only a few sexual morphs have been studied (Hyde et al. 2013). A photosensitizing toxic compound named ‘cercosporin’ is responsible for Cercospora species inhabiting such a wide host range (Daub et al. 2005; Thomas et al. 2020).
Classification – Ascomycota, Dothideomycetes, Dothideomycetidae, Capnodiales, Mycosphaerellaceae
Type species – Cercospora apii Fresen., Beitr. Mykol. 3: 91 (1863)
Distribution – Worldwide
Disease symptoms – Leaf blights and spots
This disease affects the leaves, petioles, stems and peduncles of the tree. Infection and lesion formation initially occur on older leaves before progressing to newer ones. Small, brown flecks develop with a reddish border, expanding to circular spots with an ashy-grey centre. Concentric rings may be observed as individual lesions expand. This tissue becomes thin and brittle, and often drops out, leaving a ragged hole. These lesions often resemble frogeyes, giving this disease its common name. Severely affected leaves wither and die from coalescing lesions (Shane and Teng 1992; Steddom et al. 2005).
Species of Cercospora cause blights and spots on the leaves, petioles, stems and peduncles of trees. Often infection and lesion formation occurs on older leaves before progressing to newer ones. Common symptoms include small, brown lesions that develop with a reddish border, eventually expanding to larger circular or angular spots. Concentric rings may be observed as individual lesions expand. The tissue becomes thin and brittle, and often drops out, leaving a ragged hole. Severely affected leaves wither and die from coalescing lesions (Shane and Teng 1992; Steddom et al. 2005).
Hosts – Wide host range including plant genera in Amaranthaceae, Apiaceae, Asteraceae, Arecaceae, Chenopodiaceae, Convolvulaceae, Cryptogammaceae, Cucurbitaceae, Cyatheaceae, Dennstaedtiaceae, Dioscoreaceae, Euphorbiaceae, Fabaceae, Gunneraceae, Hydrangeaceae, Lamiaceae, Lygodiaceae, Musaceae, Myrtaceae, Onagraceae, Plumbaginaceae, Poaceae, Pteridaceae, Scrophulariaceae, Solanaceae, Thelypteridaceae and Urticaceae (Farr and Rossman 2020).
Cercospora apii causes leaf spot disease on celery and C. beticola on sugar beet (Braun et al. 2013; Guatimosim et al. 2016). The pathogen Cercospora cf. sigesbeckiae infects various plant families, including economically valuable crops such as soybean, causing ‘Cercospora leaf blight’, a disease characterized by leaf bronzing (Albu et al. 2016 2017). Some other species identified as causative organisms of the leaf blight are C. kikuchii and C. cf. flagellaris (Soares et al. 2015; Rezende et al. 2020). The yield losses related to Cercospora disease have been reported from Canada, China, India and other regions in the USA and South America (Almeida et al. 2005; Cai et al. 2009; Hershman 2009; Wrather et al. 2010; Geisler et al. 2013; Albu et al. 2017; Bandara et al. 2020). Cercospora is among the leading fungal pathogens that cause a severe threat to soybean, which is an important grain legume crop, by reducing seed production and quality (Arantes et al. 2020). Two notable pathogens on soybean are C. kikuchii (leaf blight and purple seed stain) and C. sojina (frogeye leaf spot) (Soares et al. 2015)
Other notable reports include Cercospora leaf spots, which are the most common and destructive of the Hibiscus diseases, often resulting in complete crop loss (Park et al. 2017) and more than 200 fungal species in association with various diseases of ‘kenaf’ (Hibiscus cannabinus) worldwide (Park et al. 2017). Key proteins and expression of genes that could inhibit the pathogen C. kikuchii in soybean (Arantes et al. 2020) have been investigated. However, based on previous reports, morphological characters, phylogeny and pathogenicity of Cercospora cf. nicotianae was identified as one of several cryptic species causing Cercospora leaf blight (Sautua et al. 2019, 2020). Thomas et al. (2020) proposed the expression of fungal cercosporin auto resistance genes and silencing of the cercosporin pathway as effective strategies to combat Cercospora diseases.
Pathogen biology, disease cycle and epidemiology
The taxa survive on undecomposed residues in soil, on weed hosts and seeds. Leaf spot disease is favoured by warm, wet weather. Severe outbreaks generally require a period of showery weather. Infection from germinating fungal spores occurs via penetration of leaf stomata by fungal hyphae. Spores spread in wind, rain, irrigation or via mechanical tools (Vereijssen 2004; Lin and Kelly 2018).
Morphological based identification and diversity
Cercospora has been widely applied to all kinds of dematiaceous hyphomycetous asexual morphs characterized by holoblastic conidiogenesis and some associated with “Mycosphaerella”-like sexual morphs (Hyde et al. 2013; Groenewald et al. 2013). Species resembling the genus type, C. penicillata, characterized by pigmented conidiophores, thickened and darkened conidiogenous loci and singly formed colourless conidia are identified as Cercospora sensu stricto (Ellis 1971, 1976). Chupp (1954) published a worldwide monograph of this group which listed 1,419 species. A vast number of studies related to Cercospora are based on morphology or confined to specific regions or hosts (Phengsintham et al. 2013a, b). Hence, more than 3,000 species of Cercospora have been described (Pollack 1987), often as a result of taxa being considered as host-specific at a genus or family level (Crous and Braun 2003; Groenewald et al. 2005). However, based on morphological features of the structure of conidiogenous loci and hila, absence or presence of pigmentation in conidiophores and conidia, Crous and Braun (2003) revised the generic circumscription of Cercospora, resulting in the reduction of the number of species to 659. A series of publications related to Cercospora and its allied genera in Mycosphaerellaceae, along with illustrations and descriptions of sexual morphs was published by Braun et al. (2013, 2014, 2015a, b, 2016).
Molecular based identification and diversity
Cercospora is monophyletic (Stewart et al. 1999; Hyde et al. 2013). Groenewald et al. (2013) provided a comprehensive phylogenetic analysis of 360 isolates which included ITS, and protein-coding genes; translation elongation factor 1-alpha (tef1), actin (act), calmodulin (cal) and histone 3 (his). This provided a basis for the identification of Cercospora species, indicating most to be host-specific (Park et al. 2017). Bakhshi et al. (2018) subjected 170 Cercospora isolates to an eight-gene analysis (tef1, act, cal, his, tub2, rpb2, gapdh) which resulted in several new clades within the C. apii, C. armoraciae, C. beticola, C. cf. flagellaris and Cercospora sp. G. complexes. The combination of tef1, cal, tub2, rpb2 and gapdh provided high phylogenetic resolution for distinguishing Cercospora species with gapdh being the gene effective in distinguishing the species complexes (Bakhshi et al. 2018). The genomes for several species – Cercospora arachidicola, C. aff. canescens, C. cf. sigesbeckiae, C. kikuchii, C. sojina and C. zeae-maydis have been published, of which C. cf. sigesbeckiae and C. sojina are important soybean pathogens (Albu et al. 2017; Sautua et al. 2019). The mating-type genes of some asexual Cercospora species have been characterised (Groenewald et al. 2013), of which C. beticola, C. zeae–maydis and C. zeina are heterothallic, while only one mating type was discovered in populations of C. apii and C. apiicola (Groenewald et al. 2006, 2010).
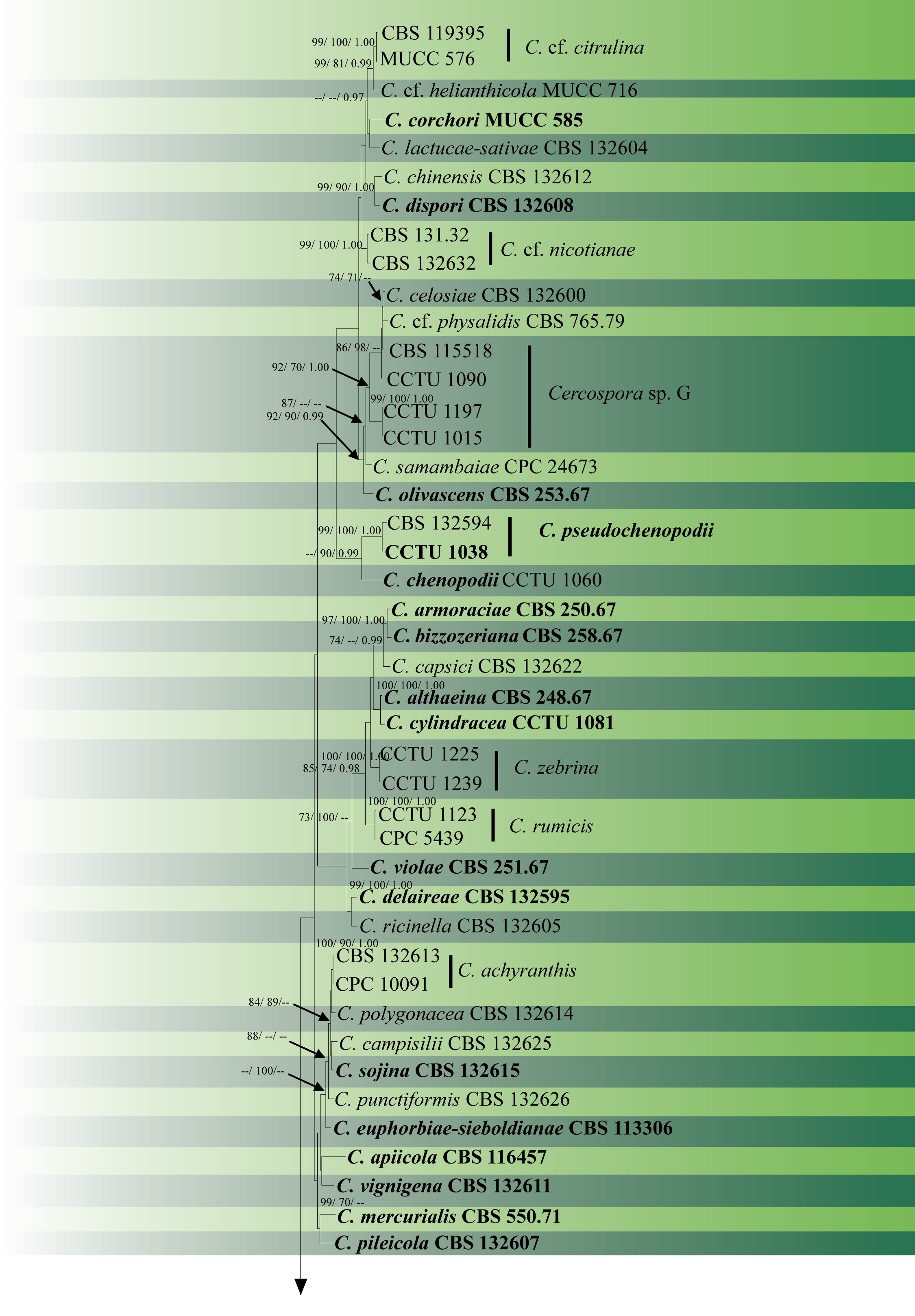
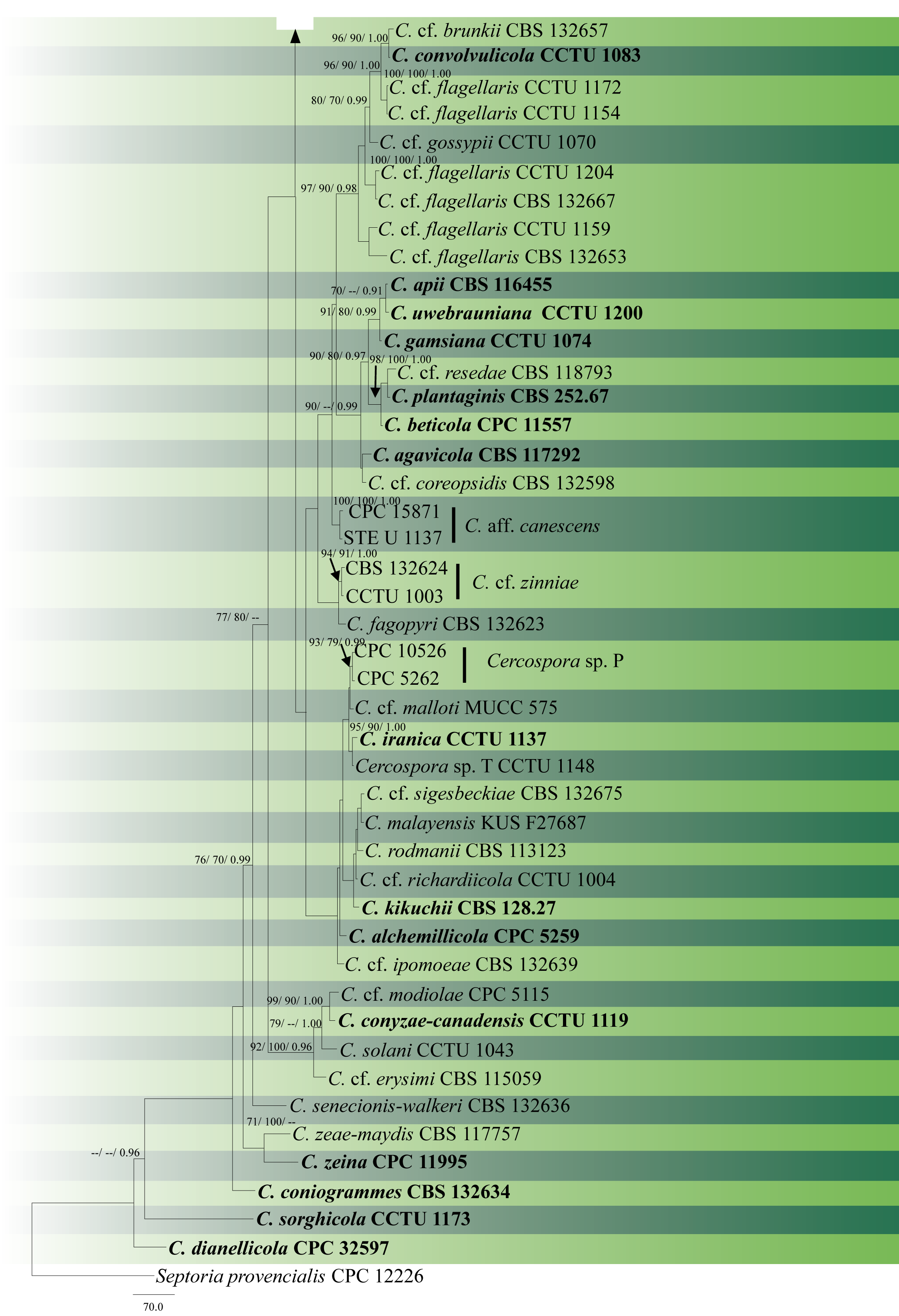
In soybean cultivation regions such as China, Latin America or the USA, C. sojina occurs as several pathotypes named as races, and their existence differs from soybean cultivar-to-cultivar (Athow et al. 1962; Yorinori and Henechin 1978; Mian et al. 2008; Gu et al. 2020). Apart from being differentiated physiologically, several molecular genetic tools such as AFLPs (Amplified Fragment Length Polymorphisms), SSR markers and SNP markers have been utilized to characterize their population diversity (Gu et al. 2020). The combination of DNA sequence data with ecology, morphological and cultural characteristics named as the Consolidated Species Concept (Quaedvlieg et al. 2014) is an effective method for delimiting Cercospora species (Groenewald et al. 2013; Bakhshi et al. 2015a, 2018). Here we provide an updated phylogenetic tree of combined ITS, tef1, act, cal, his, tub2, rpb2 and gapdh (Fig. 1).
Recommended genetic markers (genus level) – LSU, ITS
Recommended genetic markers (species level) – ITS, tef1, act, cal, his, tub2, rpb2, gapdh
Accepted number of species – There are over 3100 epithets listed in Index Fungorum (2020), however, only 134 have DNA sequence data (Table 1).
References – Braun et al. 2013, 2014, 2015a, b, 2016 (morphology), Groenewald et al. 2013 (morphology, phylogeny), Albu et al. 2017 (morphology, phylogeny), Guatimosim et al. 2016 (morphology, phylogeny), Bakhshi et al. 2015a, 2018 (morphology, phylogeny).
Table 1 DNA barcodes available for Cercospora. Ex-type/ex-epitype/ex-neotype/ex-lectotype strains are in bold and marked with an asterisk (*). Voucher strains are also in bold. Species confirmed with pathogenicity studies are marked with #.
| Species | Isolate no | ITS | tef1 | act | cal | his | tub2 | rpb2 | gapdh |
| Cercospora achyranthis | CBS 132613 | JX143523 | JX143277 | JX143031 | JX142785 | JX142539 | — | — | — |
| CPC 10091 | JX143524 | JX143278 | JX143032 | JX142786 | JX142540 | — | — | — | |
| C. agavicola# | CBS 117292* | AY647237 | AY966897 | AY966898 | AY966899 | AY966900 | — | — | — |
| C. alchemillicola# | CPC 5259* | JX143525 | JX143279 | JX143033 | JX142787 | JX142541 | — | — | — |
| C. althaeina | CBS 248.67* | JX143530 | JX143284 | JX143038 | JX142792 | JX142546 | MH496340 | — | MH496170 |
| C. apii# | CBS 116455* | AY840519 | AY840486 | AY840450 | AY840417 | AY840384 | MH496343 | — | MH496173 |
| C. apiicola# | CBS 116457* | AY840536 | AY840503 | AY840467 | AY840434 | AY840401 | — | — | — |
| C. armoraciae# | CBS 250.67* | JX143545 | JX143299 | JX143053 | JX142807 | JX142561 | MH496351 | — | MH496181 |
| C. beticola# | CBS 116456* | AY840527 | AY840494 | AY840458 | AY840425 | AY840392 | MH496355 | KT216555 | MH496185 |
| C. bizzozeriana# | CBS 258.67* | JX143546 | JX143300 | JX143054 | JX142808 | JX142562 | MH496368 | — | MH496198 |
| C. cf. brunkii | CBS 132657 | JX143559 | JX143313 | JX143067 | JX142821 | JX142575 | — | — | — |
| C. campi-silii | CBS 132625 | JX143561 | JX143315 | JX143069 | JX142823 | JX142577 | — | — | — |
| C. canescens# | CPC 15871 | JX143567 | JX143321 | JX143075 | JX142829 | JX142583 | — | — | — |
| CBS 111133 | AY260065 | DQ835084 | DQ835103 | DQ835130 | DQ835157 | — | — | — | |
| C. capsici# | CBS 132622 | JX143568 | JX143323 | JX143077 | JX142831 | JX142585 | — | — | — |
| C. celosiae | CBS 132600 | JX143570 | JX143326 | JX143080 | JX142834 | – | — | — | — |
| C. chenopodii# | CBS 132594* | JX143572 | JX143328 | JX143082 | JX142836 | JX142590 | — | — | — |
| CCTU 1060 | KJ886438 | KJ886277 | KJ885955 | KJ885794 | KJ886116 | MH496371 | MH511862 | MH496201 | |
| C. chinensis | CBS 132612 | JX143578 | JX143334 | JX143088 | JX142842 | JX142596 | — | — | — |
| C. citrullina# | CBS 119395 | EU514222 | JX143335 | JX143089 | JX142843 | – | — | — | — |
| MUCC 576 | JX143579 | JX143337 | JX143091 | JX142845 | – | — | — | — | |
| C. coniogrammes | CBS 132634* | JX143583 | JX143341 | JX143095 | JX142849 | JX142603 | — | — | — |
| C. convolvulicola | CCTU 1083* | KJ886441 | KJ886280 | KJ885958 | KJ885797 | KJ886119 | MH496374 | MH511865 | MH496204 |
| C. conyzae-canadensis | CCTU 1119* | KJ886445 | KJ886284 | KJ885962 | KJ885801 | KJ886123 | MH496377 | MH511868 | MH496207 |
| C. corchori | MUCC 585* | JX143584 | JX143342 | JX143096 | JX142850 | JX142604 | — | — | — |
| C. cf. coreopsidis | CBS 132598 | JX143585 | JX143343 | JX143097 | JX142851 | JX142605 | — | — | — |
| C. cylindracea | CCTU 1081* | KJ886449 | KJ886288 | KJ885966 | KJ885805 | KJ886127 | MH496381 | MH511872 | MH496211 |
| C. delaireae | CBS 13259* | JX143587 | JX143345 | JX143099 | JX142853 | JX142607 | — | — | — |
| C. dianellicola | CBS 143453* | MG386075 | – | MG674152 | MG674153 | – | — | — | — |
| C. dispori | CBS 13260* | JX143591 | JX143349 | JX143103 | JX142857 | JX142611 | — | — | — |
| C. cf. erysimi | CBS 115059 | JX143592 | JX143350 | JX143350 | JX142858 | JX142612 | — | — | — |
| C. euphorbiaesieboldianae | CBS 113306* | JX143593 | JX143351 | JX143105 | JX142859 | JX142613 | — | — | — |
| C. fagopyri | CBS 132623* | JX143594 | JX143352 | JX143106 | JX142860 | JX142614 | — | — | — |
| C. cf. nicotianae | CBS 131.32 | DQ835073 | DQ835099 | DQ835119 | DQ835146 | – | |||
| CBS 132632 | JX143631 | JX143631 | JX143144 | JX142898 | – | ||||
| C. cf. flagellaris# | CCTU 1159 | KJ886493 | KJ886332 | KJ886010 | KJ885849 | KJ886171 | MH496388 | MH511879 | MH496218 |
| CBS 132653 | JX143603 | JX143361 | JX143115 | JX142869 | JX142623 | MH496390 | MH511881 | MH496220 | |
| CCTU 1204 | KJ886505 | KJ886344 | KJ886022 | KJ885861 | KJ886183 | MH496399 | MH511890 | MH496229 | |
| CBS 132667 | JX143604 | JX143362 | JX143116 | JX142870 | JX142624 | MH496401 | MH511892 | MH496231 | |
| CCTU 1172 | KJ886501 | KJ886340 | KJ886018 | KJ885857 | KJ886179 | MH496409 | MH511900 | MH496239 | |
| CCTU 1154 | KJ886489 | KJ886328 | KJ886006 | KJ885845 | KJ886167 | MH496410 | MH511901 | MH496240 | |
| C. gamsiana | CCTU 1074* | KJ886426 | KJ886265 | KJ885943 | KJ885782 | KJ886104 | MH496446 | MH511937 | MH496276 |
| C. cf. gossypii | CCTU 1070 | KJ886467 | KJ886306 | KJ885984 | KJ885823 | KJ886145 | MH496452 | MH511943 | MH496282 |
| C. helianthicola# | MUCC 716 | JX143615 | JX143374 | JX143128 | JX142882 | JX142636 | — | — | — |
| C. ipomoeae# | CBS 132639 | JX143616 | JX143375 | JX143129 | JX142883 | JX142637 | — | — | — |
| C. iranica | CCTU 1137* | KJ886513 | KJ886352 | KJ886030 | KJ885869 | KJ886191 | MH496455 | MH511946 | MH496285 |
| C. kikuchii | CBS 128.27* | DQ835070 | DQ835088 | DQ835107 | DQ835134 | DQ835161 | — | — | — |
| C. lactucae-sativae | CBS 132604 | JX143621 | JX143380 | JX143134 | JX142888 | JX142642 | — | — | — |
| C. malayensis | KUS-F27687 | KR400012 | KY082663 | KY082664 | KY082665 | KY082666 | — | — | — |
| C. cf. malloti | MUCC 575 | JX143625 | JX143384 | JX143138 | JX142892 | JX142646 | — | — | — |
| C. mercurialis | CBS 550.71* | JX143628 | JX143628 | JX143141 | JX142895 | JX142649 | — | — | — |
| C. cf. modiolae | CPC 5115 | JX143630 | JX143389 | JX143143 | JXJX142897 | JX142651 | — | — | — |
| C. cf. nicotianae | CBS 131.32 | DQ835073 | DQ835099 | DQ835119 | DQ835146 | DQ835173 | — | — | — |
| CBS 132632 | JX143631 | JX143390 | JX143144 | JX142898 | JX142652 | — | — | — | |
| C. olivascens | CBS 253.67* | JX143632 | JX143391 | JX143145 | JX142899 | JX142653 | — | — | — |
| C. cf. physalidis# | CBS 765.79 | JX143633 | JX143392 | JX143146 | JX142900 | – | — | — | — |
| C. pileicola | CBS 132607* | JX143634 | JX143393 | JX143147 | JX142901 | JX142655 | — | — | — |
| C. plantaginis | CBS 252.67* | DQ233318 | DQ233342 | DQ233368 | DQ233394 | DQ233420 | MH496461 | — | MH496291 |
| C. polygonacea | CBS 132614 | JX143637 | JX143396 | JX143150 | JX142904 | JX142658 | — | — | — |
| C. pseudochenopodii | CBS 136022* | KJ886516 | KJ886355 | KJ886033 | KJ885872 | KJ886194 | MH496464 | MH511954 | MH496294 |
| C. punctiformis | CBS 132626 | JX143638 | JX143397 | JX143151 | JX142905 | JX142659 | — | — | — |
| C. cf. resedae | CBS 118793 | JX143639 | JX143398 | JX143152 | JX142906 | JX142660 | — | — | — |
| C. cf. richardiicola# | CBS 132627 | JX143640 | JX143399 | JX143153 | JX142907 | JX142661 | MH496465 | MH511955 | MH496295 |
| C. ricinella# | CBS 132605 | JX143646 | JX143405 | JX143159 | JX142913 | JX142667 | — | — | — |
| C. rodmanii | CBS 113123 | DQ835076 | AF146136 | DQ835122 | DQ835149 | DQ835176 | — | — | — |
| C. rumicis | CCTU 1123 | KJ886521 | KJ886360 | KJ886038 | KJ885877 | KJ886199 | MH496466 | MH511956 | MH496296 |
| C. rumicis | CPC 5439 | JX143648 | JX143407 | JX143161 | JX142915 | JX142669 | — | — | — |
| C. samambaiae | CPC 24673 | KT037514 | KT037474 | KT037596 | KT037463 | KT037555 | — | — | — |
| C. senecioniswalkeri | CBS 132636 | JX143649 | JX143408 | JX143162 | JX142916 | JX142670 | — | — | — |
| C. cf. sigesbeckiae# | CBS 132675 | JX143655 | JX143414 | JX143168 | JX142922 | JX142676 | — | — | — |
| C. sojina | CBS 132615 | JX143659 | JX143419 | JX143173 | JX142927 | JX142681 | — | — | — |
| C. solani# | CCTU 1043 | KJ886523 | KJ886362 | KJ886040 | KJ885879 | KJ886201 | MH496469 | MH511959 | MH496299 |
| C. sorghicola | CCTU 1173* | KJ886525 | KJ886364 | KJ886042 | KJ885881 | KJ886203 | MH496471 | MH511961 | MH496301 |
| Cercospora sp. P | CBS 116365* | AY752141 | AY752176 | AY752204 | AY752235 | AY752266 | — | — | — |
| Cercospora sp. P | JZG-2013 | JX143714 | JX143473 | JX143227 | JX142981 | JX142735 | — | — | — |
| Cercospora sp. G | CCTU 1197 | KJ886540 | KJ886379 | KJ886057 | KJ885896 | KJ886218 | MH496472 | MH511962 | MH496302 |
| Cercospora sp. G | CCTU 1015 | KJ886528 | KJ886367 | KJ886045 | KJ885884 | KJ886206 | MH496473 | MH511963 | MH496303 |
| Cercospora sp. G | CCTU 1090 | KJ886536 | KJ886375 | KJ886053 | KJ885892 | KJ886214 | MH496476 | MH511965 | MH496306 |
| Cercospora sp. G | CBS 115518 | JX143681 | JX143441 | JX143195 | JX142949 | JX142703 | MH496480 | — | MH496310 |
| Cercospora sp. T | CCTU 1148 | KJ886541 | KJ886380 | KJ886058 | KJ885897 | KJ886219 | MH496488 | MH511976 | MH496318 |
| C. uwebaruniana | CCTU 1200* | KJ886408 | KJ886247 | KJ885925 | KJ885764 | KJ886086 | MH496489 | MH511977 | MH496319 |
| C. vignigena | CBS 132611* | JX143734 | JX143493 | JX143247 | JX143001 | JX142755 | — | — | — |
| C. violae# | CBS 251.67* | JX143737 | JX143496 | JX143250 | JX143004 | JX142758 | MH496492 | — | MH496322 |
| C. zeae-maydis# | CBS 117757* | DQ185074 | DQ185086 | DQ185098 | DQ185110 | DQ185122 | — | — | — |
| C. zebrina# | CCTU 1225 | KJ886550 | KJ886389 | KJ886067 | KJ885906 | KJ886228 | MH496495 | MH511981 | MH496325 |
| CCTU 1239 | KJ886551 | KJ886390 | KJ886068 | KJ885907 | KJ886229 | MH496504 | MH511987 | MH496334 | |
| C. zeina# | CBS 118820* | DQ185081 | DQ185093 | DQ185105 | DQ185117 | DQ185129 | — | — | — |
| C. cf. zinniae | CBS 132624 | JX143756 | JX143518 | JX143272 | JX143026 | – | — | — | — |
| CCTU 1003 | KJ886552 | KJ886391 | KJ886069 | KJ885908 | KJ886230 | MH496505 | MH511988 | MH496335 |
Fig. 1 The most parsimonious tree generated by MP analysis of combined ITS, tef1, act, cal, his, tub2, rpb2 and gapdh sequence data of Cercospora species is presented. Related sequences were obtained from previous publications and GenBank. Eighty-eight strains are included in the analysis comprising 4242 characters including gaps, of which 3044 characters are constant, 464 characters are parsimony-uninformative and 734 are parsimony-informative. The parsimony analysis of the data matrix resulted in the maximum of 84 equally most parsimonious trees with a length of 2886 steps (CI = 0.556, RI=0.700, RC = 0.389, HI = 0.444) in the first tree. The tree was rooted with Septoria provencialis (CPC 12226). Tree topology of the MP analysis was similar to the ML and BYPP analyses. ML and MP bootstrap support value ≥70% and BYPP ≥0.95 (ML/ MP/ BYPP) are shown respectively near the nodes. Ex-type strains are in bold.

No Comments