18 Sep Pseudopyricularia
Pseudopyricularia Klaubauf, M.-H. Lebrun & Crous, in Klaubaufet al., Stud. Mycol. 79: 109 (2014)
Background
Pseudopyricularia is a dematiaceous hyphomycete genus introduced by Klaubauf et al. (2014) based on the type species P. kyllingae Klaubauf, Lebrun & Crous. The genus name refers to its morphological similarity to Pyricularia. Pseudopyricularia species are plant pathogens mostly associated with sedges, but they can also occur on other plants. Pseudopyricularia taxa have been also recorded as saprobes, e.g. P. higginsii was found saprobic on dead leaves of Typha orientalis (Typhaceae) (Klaubauf et al. 2014).
Classification– Sordariomycetes, Diaporthomycetidae, Magnaporthales, Pyriculariaceae
Type species – Pseudopyricularia kyllingae Klaubauf, M.-H. Lebrun & Crous, in Klaubaufet al., Stud. Mycol. 79: 109 (2014)
Distribution – Iran, Israel, Japan, New Zealand and Philippines
Disease Symptoms – Leaf spot
The symptoms start as minute scattered angular, water-soaked translucent spots on the lower surface of leaves, which enlarge and appear on the upper surface.
Hosts – Main pathogens on Cyperaceae (Klaubauf et al. 2014). Pseudopyricularia bothriochloae was found on Bothriochloa bladhii (Poaceae) causing angular leaf spots (Marin-Felix et al. 2017), and P. iraniana can infect leaves of Juncus sp. (Pordel et al. 2017).
Morphological based identification and diversity
Pseudopyricularia species are characterized by solitary conidiophores with mostly terminal conidiogenous cells that form a rachis with several protruding, flat-tipped denticles, and obclavate, brown, guttulate, septate conidia with a truncate, slightly protruding, not darkened hilum (Klaubauf et al. 2014). Ellis (1976) considered Pyricularia higginsii (presently referred to as Pseudopyricularia higginsii) as a synonym under Dactylaria. However, it was not accepted by subsequent studies (Bussaban et al. 2005; Klaubauf et al. 2014). Some Pseudopyricularia species were formerly described in Pyricularia. Several isolates previously recognized as Pyricularia higginsii were later confirmed as a species complex which represents three related species (P. cyperi, P. kyllingae, P. higginsii) belonging to Pseudopyricularia (Klaubauf et al. 2014). Pyricularia bothriochloae was also transferred to Pseudopyricularia bothriochloae (Marin-Felix et al. 2017). Species of Pseudopyricularia can be mainly differentiated from Pyricularia sensu stricto by having short, determinate, brown conidiophores with an apical rachis with flat-tipped denticles. However, because of the similarity of conidial characters, morphological species identification of Pseudopyricularia is challenging. Conidial characters cannot be used alone as a taxonomic criterion at generic level without phylogenetic analyses (Klaubauf et al. 2014).
Molecular based identification and diversity
DNA sequence data is crucial for species identification in Pseudopyricularia and morphology similar taxa. Previous studies were mainly based on morphological identification. The order Magnaporthales previously comprised the monotypic family Magnaporthaceae which contains 13 genera and more than 100 species (Zhang et al. 2011; Illana et al. 2013; Luo and Zhang 2013; Klaubauf et al. 2014). Klaubauf et al. (2014) carried out phylogenetic analyses on Pyricularia species based on combined ITS, LSU, RPB1, ACT, CAL sequence data. The result revealed two new families, namely Ophioceraceae and Pyriculariaceae, and ten new genera, including Pseudopyricularia; three species were included in Pseudopyricularia. Crous et al. (2015) described the fourth species P. hagahagae Crous & M.J. Wingf. based on LSU sequence data. Marin-Felix et al. (2017) found that P. bothriochloae was located in the Pseudopyricularia clade in a phylogenetic tree based on ITS and LSU sequence data, thus P. bothriochloae was combined under P. bothriochloae. The latest phylogenetic study on Pseudopyricularia was carried out by Pordel et al. (2017). LSU and RPB1 sequence data revealed two new Pseudopyricularia species, P. hyrcaniana A. Pordel & M. Javan-Nikkhah and Ps. iraniana A. Pordel & M. Javan-Nikkhah. The present study reconstructs the phylogeny based on analyses of ITS, LSU, RPB1, ACT and CAL sequence data for this genus, with all the species accepted to date, and it corresponds to previous studies (Pordel et al. 2017).
Recommended genetic markers (genus level) – LSU, RPB1
Recommended genetic markers (species level) – ACT, RPB1, ITS, CAL
Accepted number of species: Seven species
References: Klaubauf et al. 2014, Pordel et al. 2017 (morphology, phylogeny).
Table Pseudopyricularia. Details of the isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an * and voucher strains are in bold.
| Species | Isolate | ITS | LSU | RPB1 | ACT | CAL |
| Macgarvieomyces borealis | CBS 461.65* | KM484854 | DQ341511 | KM485070 | KM485170 | KM485239 |
| M. juncicola | CBS 610.82 | KM484855 | KM484970 | KM485071 | KM485171 | KM485240 |
| Pseudopyricularia bothriochloae | CBS 136427* | KF777186 | KY905701 | KY905700 | ||
| P. cyperi | CBS 133595* | KM484872 | KM484990 | AB818013 | AB274453 | AB274485 |
| P. cyperi | CBS 665.79 | KM484873 | DQ341512 | KM485093 | KM485178 | KM485248 |
| P. cyperi | PH0053 | KM484874 | KM485094 | KM485179 | KM485249 | |
| P. hagahagae | CPC 25635* | KT950851 | KT950877 | KT950873 | ||
| P. higginsii | CBS 121934 | KM484875 | KM484991 | KM485095 | KM485180 | KM485250 |
| P. hyrcaniana | IRAN2758C* | KP144447 | KP144452 | KY457270 | KY457260 | |
| P. hyrcaniana | UTFC-PO11 | KP144448 | KY457266 | KY457271 | KY457261 | |
| P. hyrcaniana | UTFC-PO12 | KM207211 | KY457267 | KY457272 | KY457262 | |
| P. iraniana | IRAN 2761C* | KY457258 | KY457268 | KY457273 | KY457264 | |
| P. iraniana | UTFC-PO12 | KM207210 | KP144454 | KY457263 | ||
| P. kyllingae | CBS 133597* | KM484876 | KM484992 | KM485096 | AB274451 | AB274484 |
| P. kyllingae | PH0054 | KM484877 | KM484993 | KM485097 | KM485181 | KM485251 |
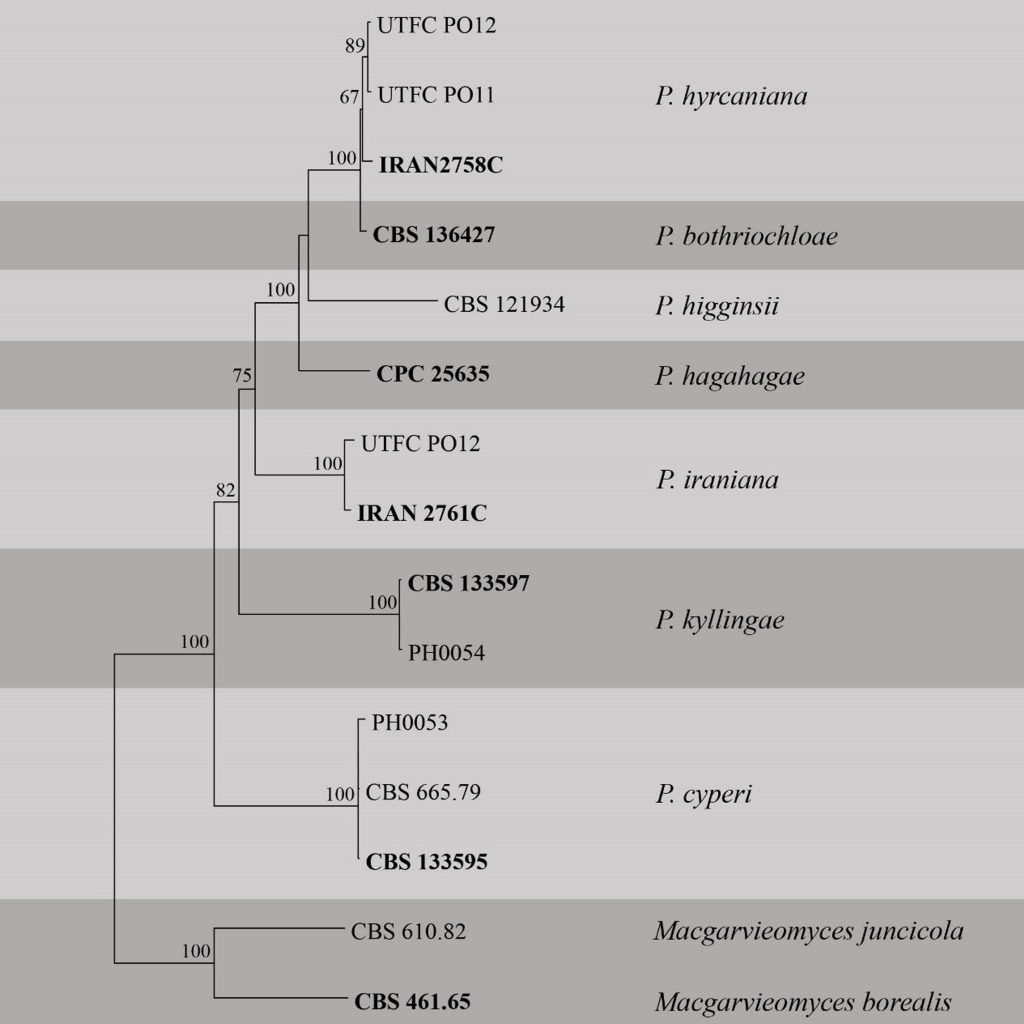
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU, RPB1, ACT and CAL sequence data of Pseudopyricularia species. Related sequences were obtained from GenBank. Fifteen strains are included in the analyses. Tree was rooted with Macgarvieomyces borealis (CBS 461.65) and M. juncicola (CBS 610.82). The best scoring RAxML tree with a final likelihood value of -10627.314729 is presented. The matrix had 771 distinct alignment patterns, with 20.13% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.244984, C = 0.283221, G = 0.271087, T = 0.200708; substitution rates AC = 1.176383, AG = 2.920484, AT = 1.126771, CG = 1.004552, CT = 6.091634, GT = 1.000000; gamma distribution shape parameter α = 1.511342. RAxML bootstrap support values ≥60% are shown respectively near the nodes. The scale bar indicates 0.02 changes. The ex-type (ex-epitype) strains are in bold.

No Comments