16 Oct Capnodium
Capnodium Mont., Annls Sci. Nat., Bot, sér. 3, 11: 233 (1849)
The genus Capnodium was introduced by Montagne (1849) to accommodate C. salicinum. Capnodium is one of the most commonly found sooty molds in gardens and landscapes (Laemmlen 2011). Capnodium has a saprobic association with sap-feeding insects in the Order Homoptera, which includes aphids, whiteflies, soft scale, mealy bugs, leafhoppers and psyllids (Barr 1987). Gavrilov-Zimin (2017) reported that the larvae and female of a new species and a new monotypic genus of legless mealybug, Orbuspedum machinator, from bamboo twigs in southern Thailand are covered with densely packed fungal hyphae of the sooty mold Capnodium sp. Herath et al. (2012) reported that a tropical sooty mold (Capnodium sp.) is known to produce antibiotics such as tetramic acid, methiosetin, and epicorazin A.
Capnodium species grow on honeydew, gradually covering the surface of the plant part affected by insects, coloring it with various shades of black. These fungi do not colonize the plant tissues or trigger symptoms. However, they alter the ability of the plant to perform photosynthesis and exchange of gases with the atmosphere. Severely affected leaves may die and fall, thereby affecting plant growth and survival. Therefore, we treat Capnodium as the main plant pathogenic group.
Classification – Dothideomycetes, Dothideomycetidae, Capnodiales, Capnodiaceae
Type species – Capnodium salicinum Mont., Annls Sci. Nat., Bot, sér. 3 11:234 (1849)
Distribution – Species of Capnodium have a wide distribution but are most common in tropical and subtropical regions (Chomnunti et al. 2014). They can be found on plants that have been previously fed upon by insects.
Disease symptoms – Dark mycelium coating surface of the host can cause chlorosis and reduce the photosynthetic ability of plants, which affects plant growth, reduces yield, and gives marketability problems (Chomnunti et al. 2014; Fig 1). In higher latitudes, Capnodium spp. are scarce during the winter; the most common being C. salicinum in the UK (Cannon et al. 1985; Royal Botanic Gardens, Kew, UK National Collection of Dried Fungi, unpublished data). Warm-temperate climates in Australia and the Mediterranean countries provide an abundance of perennial foliage on which sooty molds are able to establish themselves during the winter, and so persist from one season to the next (Fraser 1935; Reynolds and Gilbert 2005). In northern Thailand, most of the sooty mold infections are caused by Capnodium species (Chomnunti et al. 2014).
Hosts – Many plants when colonized by insects that produce honeydew. Species of Annona, Camellia, Citrus, Coffea, Chrysophyllum, Ficus, Malus, Mangifera, Olea, Populus, Prunus, Psidium, Rhododendron and Salix (Farr and Rossman 2019)
Fig Sooty molds on various host plants associated with insects. a on a hardwood tree. b on guava. c on coffee. d–f on mango.
Morphological based identification and diversity
The asexual morph forms elongated pycnidia that develop from a superficial mycelium on living plant surfaces and produce tiny, hyaline conidia on top of the pycnidia (Chomnunti et al. 2011). Persoon (1822) mentions that Fumago citri is the sooty mold but it was not well described and completed; therefore it was transferred to genus Polychaeton by Léveillé (1847). Later, Berkeley-Desmazieres (1849) transferred all species once known in the genus Fumago to Capnodium. Molecular evidence revealed that Polychaeton is an asexual stage of Capnodium, therefore, both are the same organism. According to “one fungus one name” and the Melbourne Code under Art. 57.2, Capnodium was considered for conservation as it has a larger number of epithets and is more widely used in this group of fungi, even though Polychaeton is the older name (Chomnunti et al 2011, 2014; McNeill et al. 2012; Hyde et al. 2013; Wijayawardene et al. 2014, 2017, 2018; Liu et al. 2015; Hongsanan et al. 2015).
The morphology of Capnodium species can be recognized by black mycelial growth spreading on the host surface, which produces superficial colonies with septate, dark brown hyphae and cylindrical and bitunicate asci. On host surface Capnodium species share the same ecological niche and are similar in appearance to other genera and families of sooty molds; often found with sexual and asexual states growing together and living in complex communities (Faull et al. 2002; Hughes 2003, Hughes and Seifert 2012; Chomnunti et al. 2014; Hongsanan et al 2015).
Molecular based identification and diversity
DNA sequencing data of Capnodium coffeae, C. coartatum, C. salicinum, C. coffeicola and C. dematum and eleven unidentified Capnodium spp. are available in GenBank, including sequence data for LSU, SSU and ITS (4/7/2019). Hongsanan et al. (2015) introduced a new species Capnodium coffeicola. It differs from other Capnodium species in having pycnidia with short and black stalks at the base and is swollen at the central part, and it has cylindrical to oblong conidia, but its placement is supported with phylogenetic analysis using LSU and ITS sequence data.
Sooty molds often grow in colonies of more than one species, and taxonomic descriptions thus often unknowingly combine elements of different genera and species. Identification based on morphology only is difficult as there are overlapping morphological characters among many taxa (Chomnunti et al. 2011, 2014). To achieve accurate generic and species identification and taxonomic placements, phylogenetic studies using large subunit ribosomal RNA (LSU rRNA) gene sequences and the internal transcribed spacer regions and 5.8S nrDNA gene (ITS) were performed (Crous et al. 2009; Chomnunti et al. 2011, 2014; Liu et al. 2015; Hongsanan et al. 2015).
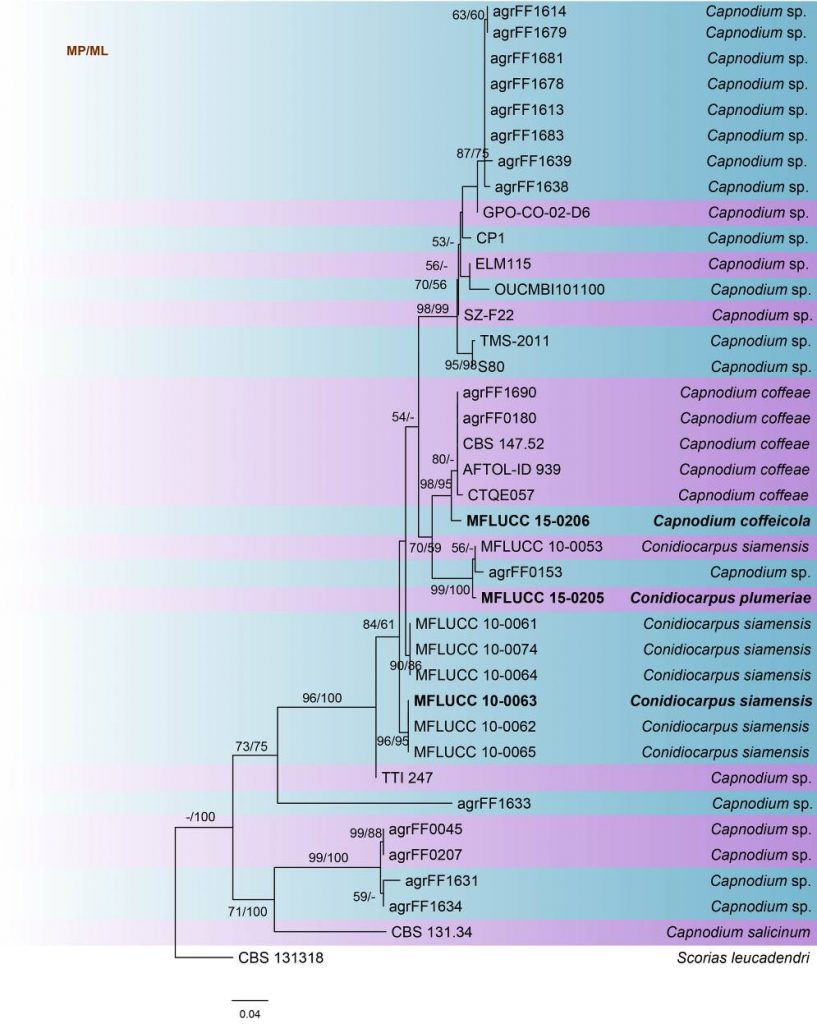
This study reconstructs the phylogeny of Capnodium based on analyses of ITS sequence data (Table 1, Fig. 2) and corresponds with previous studies (Chomnunti et al. 2011, 2014; Hongsanan et al. 2015). This can be used as a backbone tree in the identification of Capnodium species.
Fig. Morphology of Capnodium sp. a pycnidia on the host. b, d,e stalked pycnidia c mycelium network. g conical pycnidium and pycnidial wall. h ostiole surrounded by hyaline hyphae. i conidia. Scale bars: b, d–f = 100 μm, c, g = 50 μm, h, = 20 μm, i = 10 μm.
Recommended genetic markers (genus level) – LSU
Recommended genetic markers (species level) – LSU, ITS
Sequence data of LSU, SSU and ITS are available for five species of Capnodium in GenBank but none of them has complete sequence data. LSU is useful for preliminary identification at the generic level (Chomnunti et al 2011, 2014; Quaedvlieg et al 2014). Hongsanan et al. (2015) recommended the use of combined LSU and ITS sequence data to identify the species. More protein-coding gene loci should be sequenced to clarify the taxonomic problems in this genus. In the current analyses, C. cortatum was not included due to a lack of ITS sequences in GenBank. A revision of this genus is needed as it may reveal many new species. Re-sequencing of species as well as designating epitypes or representative species is also important.
The accepted number of species: There are 140 epithets in Index Fungorum (2019), however only four species have DNA sequence data.
References: Chomnunti et al 2011, 2014; Quaedvlieg et al 2014; Hongsanan et al. 2015 (morphology, phylogeny).
Table Details of Capnodium isolates used in the phylogenetic analyses. Ex-type (or ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher no | ITS |
| C. coffeae | AFTOL-ID 939 | DQ491515 |
| C. coffeae | CBS 147.52 | AJ244239 |
| Capnodium coffeae | CTQE057 | KX893384 |
| Capnodium coffeicola | MFLUCC 15-0206* | KU358921 |
| Capnodium salicinum | CBS 131.34 | AJ244240 |
| Capnodium sp. | SZ-F22 | KT443921 |
| Capnodium sp. | S80 | MH633887 |
| Capnodium sp. | CP1 | MH629975 |
| Capnodium sp. | OUCMBI101100 | HQ914834 |
| Capnodium sp. | ELM115 | KU556052 |
| Capnodium sp. | GPO-CO-02 | KC180729 |
| Capnodium sp. | agrFF1633 | HE584839 |
| Capnodium sp. | agrFF1683 | HE584838 |
| Capnodium sp. | agrFF1681 | HE584837 |
| Capnodium sp. | agrFF1679 | HE584836 |
| Capnodium sp. | agrFF1678 | HE584835 |
| Capnodium sp. | agrFF1639 | HE584834 |
| Capnodium sp. | agrFF1638 | HE584833 |
| Capnodium sp. | agrFF1614 | HE584832 |
| Capnodium sp. | agrFF1613 | HE584831 |
| Capnodium sp. | agrFF0153 | HE584830 |
| Capnodium sp. | agrFF1634 | HE584829 |
| Capnodium sp. | agrFF1631 | HE584828 |
| Capnodium sp. | agrFF0180 | HE584822 |
| Capnodium sp. | agrFF1690 | HE584823 |
| Capnodium sp. | agrFF0045 | HE584825 |
| Capnodium sp. | agrFF0207 | HE584826 |
| Capnodium sp. | TMS-2011 | HQ631045 |
| Capnodium sp. | TTI-247 | KU985278 |
| Conidiocarpus plumeriae | MFLUCC 15-0205 | KU358919 |
| Conidiocarpus siamensis | MFLUCC 10-0053 | KU358922 |
| Co. siamensis | MFLUCC 10-0061 | KU358923 |
| Co. siamensis | MFLUCC 10-0062 | KU358924 |
| Co. siamensis | MFLUCC 10-0063 | KU358925 |
| Co. siamensis | MFLUCC 10-0064 | KU358926 |
| Co. siamensis | MFLUCC 10-0065 | KU358927 |
| Co. siamensis | MFLUCC 10-0074 | KU358928 |
| Scoria leucadendri | CBS 131318 | JQ044437 |
Fig. Phylogenetic tree generated by maximum likelihood analysis of ITS sequence data of Capnodium species. Related sequences were obtained from GenBank. Thirty-eight strains are included in the analyses, which comprise 512 characters including gaps. The tree was rooted with Scoria leucadendri (CBS 131318). The tree topology of the ML analysis was similar to the MP analysis. The best scoring RAxML tree with a final likelihood value of -2398.970137 is presented. The matrix had 236 distinct alignment patterns, with 16.01% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.285338, C = 0.285338, G = 0.241464, T = 0.238894; substitution rates AC = 1.204962, AG = 1.857184, AT = 2.642904, CG = 1.319860, CT = 4.051266, GT = 1.000000; gamma distribution shape parameter α = 0.287147. The maximum parsimonious dataset consisted of constant 327, 125 parsimony-informative and 60 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 358 steps (CI = 0.735, RI = 0.875, RC = 0.630, HI = 0.265) in the first tree. RAxML and maximum parsimony bootstrap support value ≥50% are shown, respectively, near the nodes. Ex-type strains are in bold.



No Comments