16 Oct Cyttaria
Cyttaria Berk., Trans. Linn. Soc. London 19:40 (1842)
This genus is geographically restricted to South America (Argentina and Chile) and Southeastern Australasia (including Tasmania, and New Zealand) (Peterson and Pfister 2010). Cyttaria species are found in the secondary phloem and xylem, cambium and cortex of the hosts. They produce trunk and branch cankers that arise due to localized, stimulated cambial activity attributed to the presence of hyphae of Cyttaria (Wilson 1937; Gutierrez de Sanguinetti 1988). Cyttaria species are considered as weak parasites (Gamundı´ and Lederkremer 1989).
Classification – Leotiomycetes, Leotiomycetidae, Cyttariales, Cyttariaceae
Type species – Cyttaria darwinii Berk., Trans. Linn. Soc. London 19:40 (1842)
Distribution – Argentina, Australia, Chile, New Zealand, Tasmania.
Disease symptoms – Canker, galls
These species are known to cause two types of cankers: globose and longitudinal. Globose cankers arise from growth mainly in the transverse axis of the branch while longitudinal cankers arise from growth mainly along the long axis (Rawlings 1956; Gamundi 1971). The development of perennial galls on branches and stems may lead to malformation and occasional death of branches (Gadgil 1985).
Hosts – Nothofagus spp.
Morphological based identification and diversity
Ascomata of Cyttaria species are orange, pitted apothecia similar to deeply dimpled golf balls. Each fruiting body is composed of 1–200 apothecia immersed in a sterile fleshy-gelatinous stroma. Asci are 8-spored, inoperculate and amyloid. Ascospores are uninucleate, subglobose to ovoid, smooth to rugulose, at first hyaline to yellowish but later becoming pigmented (Mengoni 1986; Peterson et al 2010). There are 21 epithets listed in Index Fungorum (2019).
Molecular based identification and diversity
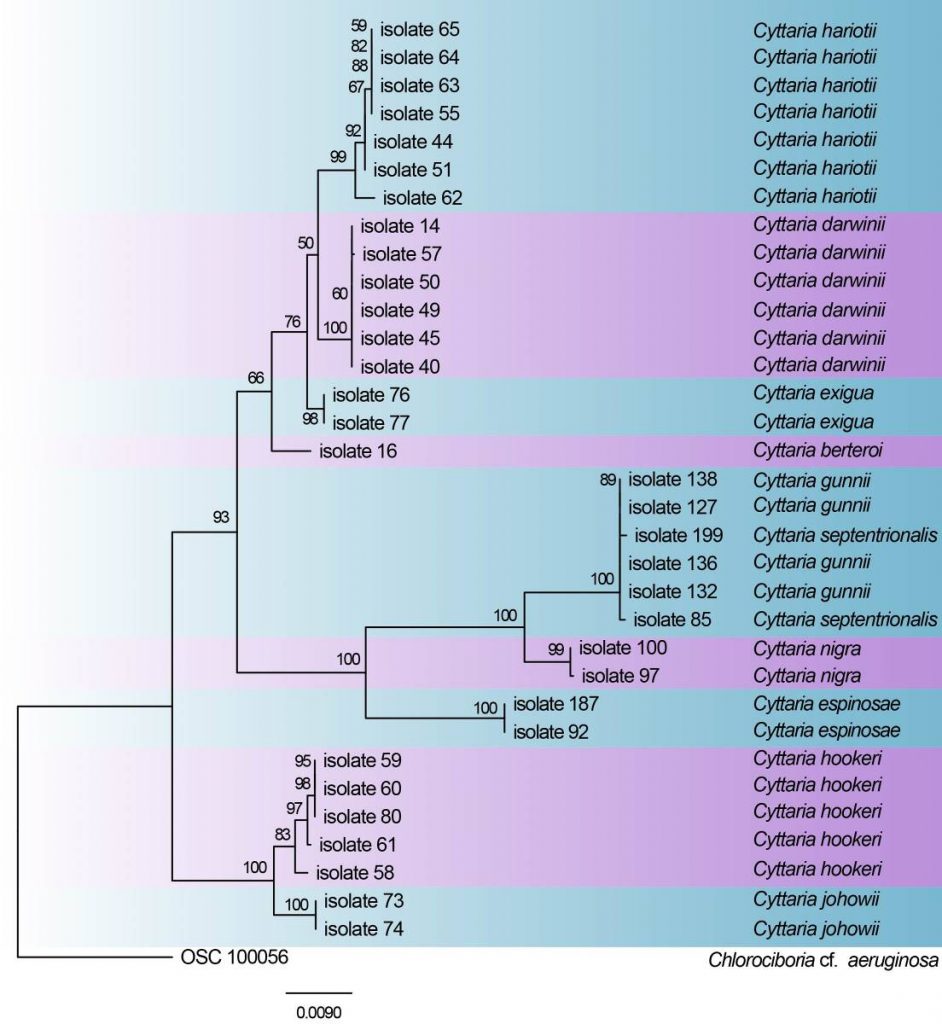
The first phylogenetic analysis which included Cyttaria was done by Gargas and Taylor (1995) showing its relationship with other discomycetes. Ekanayaka et al. (2017) Wang et al. (2006) showed its placement within Leotiomycetes using combined analysis of SSU, LSU, and 5.8S rDNA gene sequence data and then confirmed by Ekanayaka et al. (2017). Peterson and Pfister (2010) did large scale phylogeny for Cyttaria including all accepted 12 species in the genus using sequence data of partial nucSSU, nucLSU, and mitSSU rRNA, as well as TEF1. They found Cyttaria to be a strongly supported clade and suggested a close relationship between Cyttaria and some members of the Helotiales (Cordierites, Encoelia, Ionomidotis, and Chlorociboria) (Peterson and Pfister 2010). The present study reconstructs the phylogeny of Cyttaria based on analyses of a combined LSU, SSU and mtSSU sequence data (Table 5, Fig. 10). The phylogenetic tree is updated with recently introduced Cyttaria species and corresponds to previous studies (Feng et al. 2014; Gao et al. 2015).
Recommended genetic markers (genus level) – ITS, LSU
Recommended genetic markers (species level) – nucSSU, nucLSU, mitSSU rRNA, and tef1
Combined nucSSU, nucLSU, mitSSU rRNA, and tef1 can resolve almost all species of Cyttaria currently known from sequence data (Peterson et al 2010).
The accepted number of species: There are 21 epithets in Index Fungorum (2019). However, 12 species have molecular data and are treated as accepted.
References: Mengoni 1986, Peterson et al 2010 (morphology); Peterson and Pfister (2010), Ekanayaka et al. 2017 (morphology, phylogeny).
Table Details of Cyttaria isolates used in the phylogenetic analyses. Ex-type (or ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher no | LSU | SSU | mtSSU |
| C. berteroi | isolate 16 | EU107205 | EU107178 | EU10723 |
| C. darwinii | isolate 40 | EU107207 | EU107180 | EU107236 |
| C. darwinii | isolate 14 | EU107208 | EU107181 | — |
| C. darwinii | isolate 57 | EU107206 | EU107179 | EU107235 |
| C. darwinii | isolate 45 | EU107209 | — | — |
| C. darwinii | isolate 50 | EU107211 | — | — |
| C. darwinii | isolate 49 | EU107210 | — | — |
| C. espinosae | isolate 187 | — | EU107183 | EU107238 |
| C. espinosae | isolate 92 | EU107212 | EU107182 | EU107237 |
| C. exigua | isolate 77 | EU107214 | EU107185 | EU107240 |
| C. exigua | isolate 76 | EU107213 | EU107184 | EU107239 |
| C. gunnii | isolate 138 | — | EU107189 | EU107242 |
| C. gunnii | isolate 127 | EU107215 | EU107186 | EU107241 |
| C. gunnii | isolate 136 | — | EU107188 | — |
| C. gunnii | isolate 132 | — | EU107187 | — |
| C. hariotii | isolate 44 | EU107217 | EU107194 | EU107245 |
| C. hariotii | isolate 55 | EU107218 | EU107195 | EU107246 |
| C. hariotii | isolate 65 | EU107223 | — | — |
| C. hariotii | isolate 64 | EU107222 | — | — |
| C. hariotii | isolate 63 | EU107221 | — | — |
| C. hariotii | isolate 62 | EU107220 | — | — |
| C. hariotii | isolate 51 | EU107219 | — | — |
| C. hookeri | isolate 60 | EU107227 | — | — |
| C. hookeri | isolate 59 | EU107226 | — | — |
| C. hookeri | isolate 80 | EU107228 | — | — |
| C. hookeri | isolate 61 | EU107225 | EU107197 | — |
| C. hookeri | isolate 58 | EU107224 | EU107196 | — |
| C. johowii | isolate 73 | EU107229 | EU107198 | — |
| C. johowii | isolate 74 | EU107230 | EU107199 | — |
| C. nigra | isolate 100 | EU107232 | EU107201 | EU107248 |
| C. nigra | isolate 97 | EU107231 | EU107200 | EU107247 |
| C. septentrionalis | isolate 199 | — | EU107203 | EU107249 |
| C. septentrionalis | isolate 85 | — | EU107202 | — |
Fig. Phylogram generated from RAxML analysis based on combined sequences of LSU, SSU and mtSSU sequences of all the accepted species of Cyttaria. Related sequences were obtained from GenBank. Thirty-four strains are included in the analyses, which comprise 3480 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted with Chlorociboria cf. aeruginosa (OSC 100056). The best scoring RAxML tree with a final likelihood value of -7505.900855 is presented. The matrix had 360 distinct alignment patterns, with 39.17% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.256, C = 0.214, G = 0.280, T = 0.250; substitution rates AC = 1.190197, AG = 1.207782, AT = 0.373130, CG = 0.681125, CT = 3.724394, GT = 1.000000; gamma distribution shape parameter α = 0.020000. RAxML and maximum parsimony bootstrap support value ≥50% (BT) are shown respectively near the nodes.

No Comments