23 Oct Diplodia
Diplodia
Background
Species of Diplodia (Botryosphaeriaceae) are endophytes, pathogens, or saprobes associated with cankers, dieback and fruit rot (Crous et al. 2006; Slippers and Wingfield 2007) in a wide range of hosts of agricultural and forestry importance (Farr and Rossman 2014). Cryptic speciation is common in the genus Diplodia, which makes species identification difficult if only based on morphological characters (Phillips et al. 2012, 2013). Denman et al. (2000) suggested that Lasiodiplodia could be a synonym of Diplodia, however recent studies accepted them as distinct genera (Pavlic et al. 2004; Burgess et al. 2006; Damm et al. 2007; Alves et al. 2008).
The genus Diplodia was introduced by Montagne (1834) with concepts altering over the years and has been regarded as including species with dark brown, 1-septate conidia (Phillips et al. 2005). Diplodia is defined by having uni or multilocular conidiomata lined with conidiogenous cells that form hyaline, aseptate, thick-walled conidia at their tips (Phillips et al. 2005). Diplodia mutila is the type species of Diplodia (Montagne 1834; Fries 1849), however, there are no living cultures linked to the holotype. As this has severely hampered studies on taxonomy and phylogeny of Diplodia, Alves et al. (2004) provided a detailed description of this species based on one isolate from grapevines in Portugal (CBS 112553).
Species identification and numbers
Diplodia is a large genus and a search in MycoBank (2014) revealed 1,317 names. Species in Diplodia were described, often based on host association, which later resulted in a proliferation of species names. According to Slippers et al. (2004d), the host is not of primary importance in species differentiation, thus, many of the names in Diplodia are likely to be synonyms.
Based on DNA sequence data (single or multimarker) and minor differences in conidial morphology, there are currently about 20 Diplodia species (de Wet et al. 2003; Alves et al. 2004, 2006; Gure et al. 2005; Damm et al. 2007; Lazzizera et al. 2008; Pérez et al. 2010; Jami et al. 2012; Phillips et al. 2012, 2013; Linaldeddu et al. 2013; Lynch et al. 2013). The phylogenetic analysis was performed based on up to date holotype or ex-epitype sequence data available in GenBank (Table).
Table Diplodia. Details of the isolates used in the phylogenetic tree
| Species | Isolate no. | Host | GenBank | ||
| ITS | TEF | β-tubulin | |||
| Diplodia africana | CBS 120835* | Prunus persica | EF445343 | EF445382 | – |
| D. agrifolia | CBS 132777* | Quercus agrifolia | JN693507 | JQ517317 | JQ411459 |
| D. alatafructa | CBS 124931* | Pterocarpus angolensis | FJ888460 | FJ888444 | – |
| D. allocellula | CBS 130408* | Acacia karroo | JQ239397 | JQ239384 | JQ239378 |
| D. bulgarica | CBS 124254* | Malus sylvestris | GQ923853 | GQ923821 | – |
| D. corticola | CBS 112549* | Quercus suber | AY259100 | AY573227 | DQ458853 |
| D. cupressi | CBS 168.87* | Cupressus sempervirens | DQ458893 | DQ458878 | DQ458861 |
| D. fraxini | CBS 136010* | Fraxinus angustifolia | KF307700 | KF318747 | – |
| D. intermedia | CBS 124462* | Malus sylvestris | GQ923858 | GQ923826 | – |
| D. malorum | CBS 124130* | Malus sylvestris | GQ923865 | GQ923833 | – |
| D. mutila | CBS 112553* | Vitis vinifera | AY259093 | AY573219 | DQ458850 |
| D. olivarum | CBS 121887* | Olea europaea | EU392302 | EU392279 | HQ660079 |
| D. sapinea | CBS 393.84* | Pinus nigra | DQ458895 | DQ458880 | – |
| D. pseudoseriata | CBS 124906* | Blepharocalyx salicifolius | EU080927 | EU863181 | – |
| D. quercivora | CBS 133852* | Quercus canariensis | JX894205 | JX894229 | – |
| D. rosulata | CBS 116470* | Prunus africana | EU430265 | EU430267 | EU673132 |
| D. scrobiculata | CBS 109944* | Pinus greggii | DQ458899 | DQ458884 | AY624258 |
| D. seriata | CBS 112555* | Vitis vinifera | AY259093 | AY573219 | DQ458856 |
| D. subglobosa | CBS 124133* | Lonicera nigra | GQ923856 | GQ923824 | – |
| D. tsugae | CBS 418.64* | Tsuga heterophylla | DQ458888 | DQ458873 | DQ458855 |
| Lasiodiplodia theobromae | CBS 164.96* | Fruit along coral reef coast | AY640255 | AY640258 | EU673110 |
Ex-type (ex-epitype) strains are bolded and marked with an * and voucher stains are bolded
Molecular phylogeny
Studies on the taxonomy and phylogeny of Diplodia were hampered by a lack of an ex-type culture linked to the generic type, D. mutila. A collection of D. mutila from Populus with an ex-type culture was designated as epitype by Alves et al. (2014). They obtained a large collection of Diplodia strains from ash and other woody hosts showing V-shaped cankers and branch dieback. These strains were identified based on morphological characters and DNA sequence data. Since 2003 several new species have been described in Diplodia and these species were recognized mainly from DNA sequence data. Diplodia scrobiculata was differentiated from D. sapinea on the basis of multiple gene genealogies inferred from six protein-coding genes and six microsatellite loci (de Wet et al. 2003). Diplodia africana (Damm et al. 2007), D. olivarum (Lazzizera et al. 2008) and D. cupressi (Alves et al. 2006) have been differentiated from D. mutila on the basis of formation of distinct clades in phylogenies based on ITS and TEF sequence data and due to their unique conidial morphology (Phillips et al. 2012).
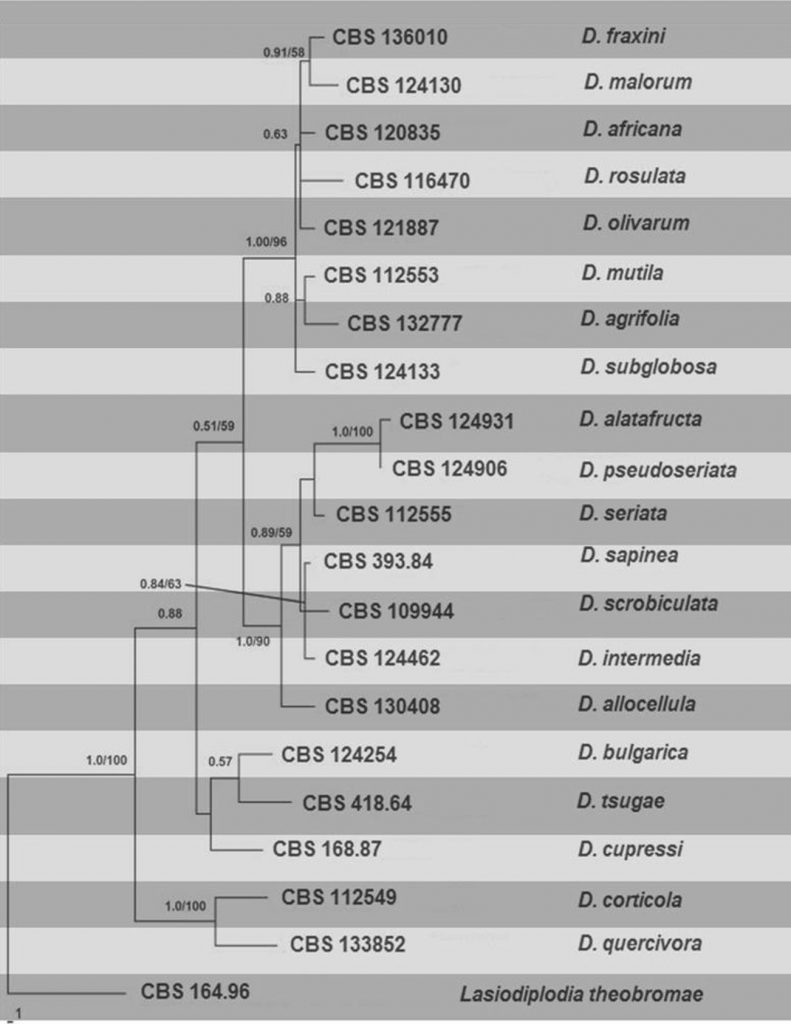
Combined morphological and phylogenetic analyses of DNA sequence data from ITS and TEF (Alves et al. 2014) showed that the Fraxinus isolates from Italy, Netherlands, Portugal, and Spain belong to three distinct species namely Diplodia fraxini, D. mutila, and D. subglobosa. The phylogenetic tree constructed with holotype or ex-epitype sequences is presented in Fig.
Fig. Phylogramgenerated from parsimony analysis based on combined ITS, TEF and β- tubulin sequenced data of Diplodia. Parsimony bootstrap support values and Bayesian posterior probabilities greater than 50 % are indicated above the nodes. The ex-type (ex-epitype) and voucher strains are in bold. The tree is rooted with Lasiodiplodia theobromae CBS 164.96.
Recommended genetic markers
- LSU and SSU– generic level
- ITS, TEF and β-tubulin– species level
ITS, TEF and β-tubulin are the common genetic markers used in the identification of Diplodia species. Combined ITS and TEF genes provide a satisfactory resolution for resolving species.

No Comments