16 Oct Entoleuca
Entoleuca Syd., Annls mycol. 20(3/4):186 (1922)
The genus Entoleuca Syd. (Xylariaceae) consists of saprobic and plant pathogenic species distributed in Europe. Entoleuca mammata causes canker diseases (commonly known as Hypoxylon canker) on Malus sp. (Rosaceae), Populus sp., Salix sp. (Salicaceae) and Sorbus sp. (Rosaceae) (Shaw 1973; Callan 1998; Kasanen et al. 2004; Eriksson 2014) and also occurs as a saprobe on decaying tree trunks. The species are distributed in terrestrial habitats in temperate regions. The genus is characterized by its known sexual morph. It is characterized by partially embedded solitary or aggregated orbicular stroma, that has a whitish surface when young and dark surface at maturity, papillate ostiole; multiple, monostichous and embedded ascomata in stromata; 8-spored, unitunicate asci that are cylindrical, long pedicellate, with J+ apical ring bluing in Melzer’s reagent and uniseriate, unicellular, ellipsoidal inequilateral, brown, with straight to oblique germ slit ascospores (Rogers and Ju 1996; Daranagama et al. 2018). Daranagama et al. (2018) provided an identification key with an emphasis on the coarsely papillate ostiole in Entoleuca.
Sydow and Petrak (1922) introduced the genus with E. callimorpha as the type species. Until 1994, Hypoxylon mammatum was considered a similar taxon to E. callimorpha. However, Læssøe and Spooner (1994) and Læssøe (1994) treated H. mammatum as a separate, synonym to Rosellinia. Based on these taxonomic confusions, Rogers and Ju (1996) revised the type, authentic and other specimens and re-established the genus Entoleuca.
Classification – Sordariomycetes, Xylariomycetidae, Xylariales, Xylariaceae
Type species – Entoleuca callimorpha Syd., in Sydow & Petrak, Annls mycol. 20(3/4):186 (1922)
Distribution – Austria, Canada, Poland, Sweden, USA
Disease symptoms – Canker
Symptoms may vary on the stage of disease development. Young cankers appear as slightly sunken, yellowish-orange areas with irregular margins. Later, the outer-most bark within the canker breaks out in blisters exposing a powdery grey mat of fungal tissue and conidia. Then the patches of bark start to flake off making the canker rough and black in the center. Advancing margins of the enlarging cankers become yellowish orange (Ostry 2013).
Hosts – Known from Malus sylvestris, Populus sp., Salix sp. and Sorbus aucuparia.
Morphological based identification and diversity
Currently, the genus comprises three species: E. callimorpha, E. ellisii and E. mammatum (Sydow and Petrak 1922; Rogers and Ju 1996; Ju et al. 2004; Index Fungorum 2019). Due to the presence of clear papillate ostioles, they have been distinguished from closely related genera such as Amphirosellinia, Nemania, and Rosellinia. Molecular data are only available for E. mammatum, which is the most important species in the genus as a pathogen. Rogers and Ju (1996) observed that there are no distinguishing morphological differences among E. mammatum isolates from different hosts. However, there is a high polymorphism, but no major phylogenetic differences among the isolates from Europe (Kasanen et al. 2004). Ju et al. (2004) introduced E. ellisii based on characterizations of ascospore and germ slit. Therefore, the combination of morphological and phylogenetic analyses is needed for species delimitation of Entoleuca.
Molecular based identification and diversity
Several recent studies have focused on the molecular phylogeny of Entoleuca, especially E. mammatum. Phylogenetic based population studies revealed that higher polymorphism occurs in North American than in Europe (Kasanen et al. 2004). The sterile mycelia associated with Pinus tabulaeformis and its ITS-based phylogenetic analyses revealed that the genus Entoleuca clusters in Xylariaceae and is closely related to Nemania (Guo et al. 2003). Daranagama et al. (2018) revisited the family Xylariaceae and due to the morphological differences and conidial state characters, they suggested that it is useful to maintain the taxa as distinct genera. Daranagama et al. (2018) and Wendt et al. (2018) conducted multi-gene phylogenetic analyses using ITS, LSU, RPB2 and TUB2 and revealed that E. mammatum clusters with Rosellinia corticium with high support. Due to the lack of molecular data from other species and other gene regions, it is difficult to place Entoleuca in an appropriate family.
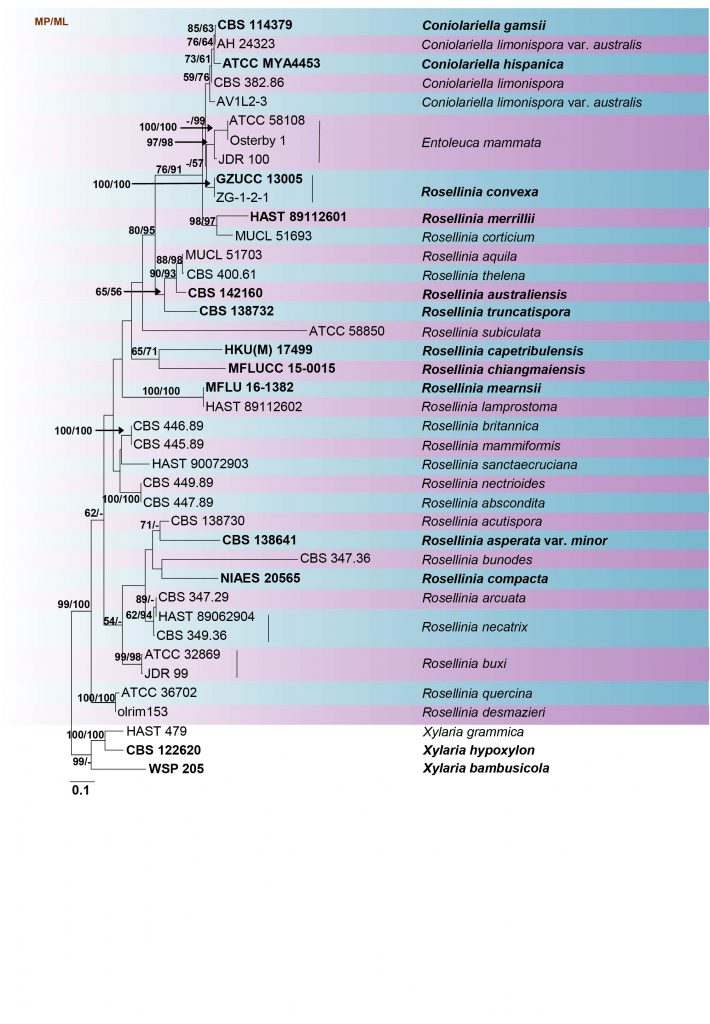
In this study, we included the available sequences of Entoleuca in the analysis done for Rosellinia (Table 7, Fig 13).
Recommended genetic markers (genus level) – LSU and ITS
Recommended genetic markers (species level) – RPB2 and TUB2
Combined LSU, ITS, RPB2, and TUB2 provide a satisfactory resolution for resolving species.
The accepted number of species: Three species
References: Sydow and Petrak 1922; Rogers and Ju 1996; Ju et al. 2004 (morphology), Daranagama et al. 2018 (morphology, phylogeny).
Table Details of the Entoleuca and Rosellinia isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher no | ITS | LSU | RPB2 |
| Coniolariella gamsii | CBS 114379* | GU553325 | GU553329 | N/A |
| C. hispanica | ATCC MYA4453* | GU553323 | GU553353 | N/A |
| C. limonispora | CBS 382.86 | KF719199 | KF719211 | N/A |
| C. limonispora var. australis | AV1L2-3 | KP101193 | N/A | N/A |
| C. limonispora var. australis | AH24323 | AY908997 | N/A | N/A |
| Entoleuca mammata | JDR 100 | GU300072 | N/A | GQ844782 |
| E. mammata | ATCC 58108 | AF201713 | N/A | N/A |
| E. mammata | Osterby 1 | AF176983 | N/A | N/A |
| Rosellinia abscondita | CBS 447.89 | FJ175180 | KF719208 | N/A |
| R. Aquila | MUCL 51703 | KY610392 | KY610460 | KY624285 |
| R. arcuate | CBS347.29 | AB017660 | N/A | N/A |
| R. asperata var. minor | CBS 138641* | KY941107 | N/A | N/A |
| R. australiensis | CBS 142160* | KY979742 | KY979797 | N/A |
| R. Britannica | CBS 446.89 | FJ175182 | KF719209 | N/A |
| R. bunodes | CBS 347.36 | AB609598 | KF719205 | N/A |
| R. buxi | JDR 99 | GU300070 | N/A | GQ844780 |
| R. buxi | ATCC 32869 | AY909000 | EF489467 | N/A |
| R. capetribulensis | HKU(M) 17499* | AY862570 | N/A | N/A |
| R. acutispora | CBS 138730 | KY941108 | N/A | N/A |
| R. chiangmaiensis | MFLU 15-3524* | KU246226 | KU246227 | N/A |
| R. compacta | NIAES:20565* | AB430457 | N/A | N/A |
| R. convexa | GZUCC 13005* | KF614036 | N/A | KP876561 |
| R. convexa | ZG-1-2-1 | KR822145 | N/A | N/A |
| R. corticium | MUCL 51693 | KC477236 | KY610461 | KY624229 |
| R. desmazieri | olrim153 | AY805591 | N/A | N/A |
| R. lamprostoma | HAST 89112602 | EF026118 | N/A | GQ844778 |
| R. mammiformis | CBS 445.89 | KF719200 | KF719212 | N/A |
| R. mearnsii | MFLU 16-1382* | KY514059 | KY514060 | KY514061 |
| R. merrillii | HAST 89112601* | GU300071 | N/A | GQ844781 |
| R. necatrix | HAST 89062904 | EF026117 | AY083824 | GQ844779 |
| R. necatrix | CBS 349.36 | AY909001 | KF719204 | N/A |
| R. nectrioides | CBS 449.89 | FJ175181 | KF719213 | N/A |
| R. quercina | ATCC 36702 | AB017661 | N/A | N/A |
| R. sanctaecruciana | HAST 90072903 | GU292824 | N/A | GQ844777 |
| R. subiculata | ATCC 58850 | AY909002 | EF489468 | N/A |
| R. thelena | CBS 400.61 | KF719202 | KF719215 | N/A |
| R. truncatispora | CBS 138732* | KY941109 | N/A | N/A |
| Xylaria bambusicola | WSP 205* | EF026123 | AB376825 | GQ844802 |
| X. grammica | HAST 479 | GU300097 | N/A | GQ844813 |
| X. Hypoxylon | CBS 122620* | AM993141 | KM186301 | KM186302 |
Fig Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU and RPB2 sequence data of Entoleuca and Rosellinia species. Related sequences were obtained from GenBank. Forty strains are included in the analyses, which comprise 2336 characters including gaps. Single gene analyses were carried out and compared with each species, to compare the topology of the tree and clade stability. The tree was rooted in Xylaria bambusicola (WSP 205), X. grammica (HAST 479) and X. hypoxylon (CBS 122620). The tree topology of the ML analysis was similar to the MP and BI. The best scoring RAxML tree with a final likelihood value of -12521.202450 is presented. The matrix had 793 distinct alignment patterns, with 51.67% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.241655, C = 0.263843, G = 0.260086, T = 0.234416; substitution rates AC = 1.764265, AG = 4.237635, AT = 0.953946, CG = 1.541272, CT = 8.643978, GT = 1.000000; gamma distribution shape parameter α = 1.105097. The maximum parsimonious dataset consisted of constant 1567, 629 parsimony-informative and 140 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 2112 steps (CI = 0.545, RI = 0.668, RC = 0.364, HI = 0.455) in the first tree. RAxML and maximum parsimony bootstrap support value ≥50% are shown respectively near the nodes. Bayesian posterior probabilities ≥0.95 (BYPP) indicated as thickened black branches. Ex-type strains are in bold.

No Comments