15 Oct Lasiodiplodia
Lasiodiplodia Ellis & Everh., Bot. Gaz. 21:92 (1896)
According to Clendenin (1896), a fungus causing rot of sweet potatoes imported from Java was identified by Ellis in 1894 as a new genus and he named the fungus Lasiodiplodia tubericola. However, Ellis (1894) did not describe the fungus or publish the new genus. Clendenin (1896) provided a description of the genus and the species, attributing both to Ellis and Everhardt. Griffin and Maublanc (1909) considered that on account of the pycnidial paraphyses, Botryodiplodia theobromae, described by Patouillard (1892), was more suitably accommodated in Lasiodiplodia. Since the epithet theobromae (1892) is older than tubericola (1896), L. theobromae should be regarded as the type species of Lasiodiplodia. Neither Patouillard (1892) nor Clendenin (1896) referred to any type or other specimens of the genus or species. Pavlic et al. (2004) could not locate the types, and they could not find any specimens from the original hosts or origins. Phillips et al. (2013) designated CBS H-21411 as neotype with CBS 164.96 as culture ex-neotype.
The sexual morph has been reported for L. theobromae, but the connection with the asexual morph has not been confirmed (Phillips 2013). Sexual morphs have also been reported for L. pseudotheobromae (Tennakoon et al. 2016), L. gonubiensis (Trakunyingcharoen et al. 2015) and L. lignicola (Phillips et al. 2013) with clear evidence that connects sexual with asexual morphs.
Classification – Dothideomycetes, incertae sedis, Botryosphaeriales, Botryosphaeriaceae
Type species – Lasiodiplodia theobromae (Pat.) Griffon & Maubl., Bull. Soc. mycol. Fr. 25: 57 (1909)
Distribution – Worldwide, mostly confined to tropical and sub-tropical regions, but becoming increasingly more common in warm temperate regions.
Disease symptoms – Diebacks, cankers, fruit rots.
Hosts – Plurivorous on woody hosts.
Morphological based identification and diversity
The pigmented, 1-septate conidia with longitudinal striations together with the pycnidial paraphyses distinguish Lasiodiplodia from all other genera in Botryosphaeriaceae (Phillips et al. 2013). Striations on the conidia distinguish it from Diplodia, the conidiomata paraphyses distinguish it from Neodeightonia, which also has striate conidia. Although Barriopsis has striate conidia and paraphyses, Lasiodiplodia is unique in the Botryosphaeriaceae because striations are visible on immature, hyaline conidia. Although Phillips et al. (2013) differentiated 18 species in Lasiodiplodia on the basis of conidial morphology (especially dimensions) and morphology of the paraphyses, in reality, species in Lasiodiplodia cannot be identified with any confidence from their morphology and molecular data are necessary for definitive identifications.
Molecular based identification and diversity
Denman et al. (2000) suggested that Lasiodiplodia could be a synonym of Diplodia. When Crous et al. (2006) re-organized Botryosphaeria on the basis of LSU phylogeny they split the genus into 10 genera, but could not resolve the position of Lasiodiplodia or separate it from Diplodia. Following a multi-locus approach (SSU, ITS, LSU, tef1 and TUB2) Phillips et al. (2008) showed that Lasiodiplodia constitutes a clear phylogenetic lineage.
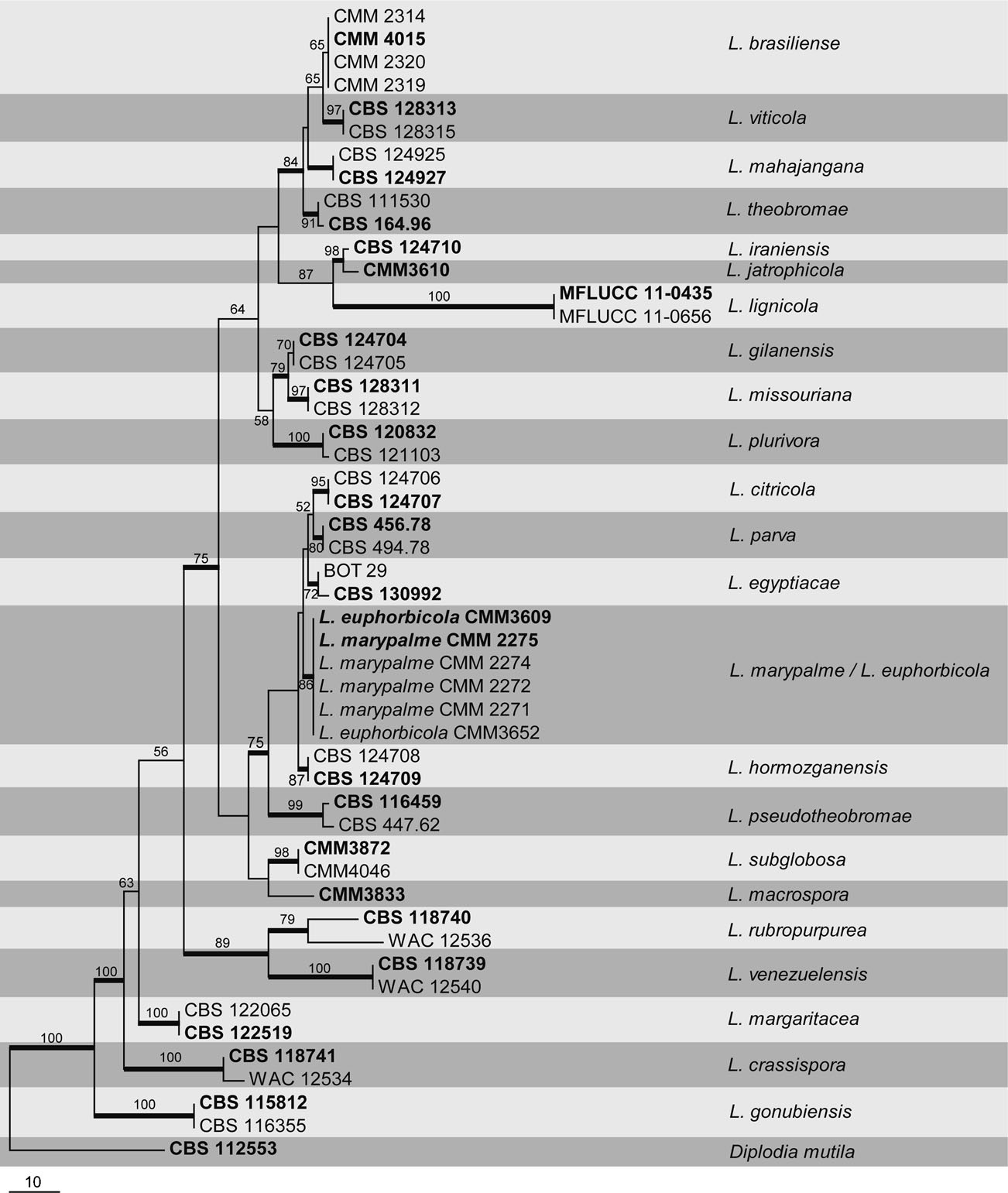
For many years, only the type species of Lasiodiplodia (L. theobromae) was mentioned in the phytopathological and mycological literature, and it was regarded as a cosmopolitan, plurivorous pathogen restricted mainly to tropical and sub-tropical regions (Punithalingam 1976, 1980). Soon after the widespread application of DNA-based phylogenies, Pavlic et al. (2004) introduced L. gonubiensis as a new species on the basis of conidial morphology and ITS sequence data. Soon after, Burgess et al. (2006) described three new species (L. crassispora, L. venezuelensis and L. rubropurpurea) from the tropics based on ITS and tef1 sequence data and morphological characters. Alves et al. (2008) also used ITS and tef1 sequence data to reveal two cryptic species in the L. theobromae complex. Over the years more species were introduced and Phillips et al. (2013) listed 18 species and Dissanayake et al. (2016) listed 31 species known from culture. Today the figure stands at 40 (Fig). Apart from L. theobromae, all species have been introduced almost entirely on the basis of DNA sequence phylogenies. Although the phylogenies were derived from analysis of multiple loci (mostly ITS, tef1 and TUB2 and sometimes RPB2) the genealogical concordance phylogenetic species recognition concept (Taylor et al. 2000) has not always been strictly applied and species have been introduced on the basis of minor differences in only one locus. The result is that some species are not well separated phylogenetically (Fig), such as L. hyalina and L. thailandica, L. chinensis, L. sterculiae, L. pseudotheobromae, L. pyriformis and L. crassispora. In a detailed study of five loci of 19 Lasiodiplodia species, Cruywagen et al. (2017) concluded that several accepted species (L. viticola, L. missouriana, L. laeliocattleyae, L. brasiliense) may, in fact, be hybrids. There has been no such study of the 16 species introduced after the work of Cruywagen et al. (2017). In view of the questionable status of several species in Lasiodiplodia, there is an urgent need to re-assess all of the species currently accepted in this genus.
Recommended genetic markers (genus level) – SSU and LSU
Recommended genetic markers (species level) – ITS, tef1, TUB2
Accepted number of species: Currently, 51 species names are listed for Lasiodiplodia in MycoBank and Index Fungorum (2019). Cultures and DNA sequences are available for 43 species, three of which have been reduced to synonymy under existing names. Thus, 40 species are currently recognised in Lasiodiplodia.
.
References: Phillips et al. 2013 (morphology, phylogeny, distribution, hosts); Dissanayake et al. 2016 (species).
Table Details of the Lasiodiplodia isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold.
| Species | Isolate/Voucher No | ITS | tef1 |
| Lasiodiplodia avicenniae | CBS 139670* | KP860835 | KP860680 |
| L. avicenniarum | MFLUCC 17-2591* | MK347777 | MK340867 |
| L. brasiliense | CMM 4015* | JX464063 | JX464049 |
| L. bruguierae | CBS 139669* | KP860832 | KP860677 |
| L. caatinguensis | CMM 1325* | KT154760 | KT008006 |
| L. chinensis | CGMCC 3.18061* | KX499889 | KX499927 |
| L. chonburiensis | MFLUCC 16-0376* | MH275066 | MH412773 |
| L. cinnamomi | CFCC 51997* | MG866028 | MH236799 |
| L. citricola | CBS 124707* | GU945354 | GU945340 |
| L. crassispora | CBS 118741* | DQ103550 | EU673303 |
| L. euphorbiicola | CMM 3609* | KF234543 | KF226689 |
| L. exigua | CBS 137785* | KJ638317 | KJ638336 |
| L. gilanensis | CBS 124704* | GU945351 | GU945342 |
| L. gonubiensis | CBS 115812* | AY639595 | DQ103566 |
| L.gravistriata | CMM 4564* | KT250949 | KT250950 |
| L. hormozganensis | CBS 124709* | GU945355 | GU945343 |
| L. hyalina | CGMCC 3.17975* | KX499879 | KX499917 |
| L. iraniensis | CBS 124710* | GU945346 | GU945334 |
| L. laeliocattleyae | CBS 167.28* | KU507487 | KU507454 |
| L. lignicola | CBS 134112* | JX646797 | KU887003 |
| L. macrospora | CMM 3833* | KF234557 | KF226718 |
| L. mahajangana | CBS 124925* | FJ900595 | FJ900641 |
| L. margaritacea | CBS 122519* | EU144050 | EU144065 |
| L. mediterranea | CBS 137783* | KJ638312 | KJ638331 |
| L. missouriana | CBS 128311* | HQ288225 | HQ288267 |
| L. pandanicola | MFLUCC 16-0265* | MH275068 | MH412774 |
| L. parva | CBS 456.78* | EF622083 | EF622063 |
| L. plurivora | CBS 120832* | EF445362 | EF445395 |
| L. pontae | CMM 1277* | KT151794 | KT151791 |
| L. pseudotheobromae | CBS 116459* | EF622077 | EF622057 |
| L. pyriformis | CBS 121770* | EU101307 | EU101352 |
| L. rubropurpurea | CBS 118740* | DQ103553 | EU673304 |
| L. sterculiae | CBS 342.78* | KX464140 | KX464634 |
| L. subglobosa | CMM 3872* | KF234558 | KF226721 |
| L. swieteniae | MFLUCC 18-0244* | MK347789 | MK340870 |
| L. thailandica | CBS 138760* | KJ193637 | KJ193681 |
| L. theobromae | CBS 164.96* | AY640255 | AY640258 |
| L. venezuelensis | CBS 118739* | DQ103547 | EU673305 |
| L. viticola | CBS 128313* | HQ288227 | HQ288269 |
| L. vitis | CBS 124060* | KX464148 | KX464642 |
Fig. Phylogenetic tree generated by maximum likelihood analysis of combined ITS and tef1 sequence data of Lasiodiplodia species. Related sequences were obtained from GenBank. Forty nine strains are included in the analyses, which comprise 866 characters including gaps. The tree was rooted with Barriopsis tectonae and B. iraniana. Tree topology of the ML analysis was similar to MP and BYPP. The best scoring RAxML tree with a final likelihood value of -3733.342990 is presented. The matrix had 253 distinct alignment patterns, with 4.41% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.211797, C = 0.285190, G = 0.260783, T = 0.242230; substitution rates AC = 0.983905, AG = 3.303939, AT = 1.281593, CG = 0.950258, CT = 5.553417, GT = 1.000000; gamma distribution shape parameter α = 0.221126. RAxML bootstrap support values ≥80% are shown respectively near the nodes. Bayesian posterior probabilities ≥0.5 (BYPP) indicated as thickened black branches. Ex-type strains are in bold.

No Comments