16 Oct Neonectria
Neonectria Wollenw., Annls mycol. 15(1/2):52 (1917)
Neonectria is a cosmopolitan genus, and their asexual morphs are common in tropical and temperate regions (Chaverri et al. 2011). Neonectria species can be found on the bark of recently dead woody plants and sometimes on decaying herbaceous material (Samuels et al. 1990; Samuels and Brayford 1990, 1993, 1994; Rossman et al. 1999; Castlebury et al. 2006; Chaverri et al. 2011). Some species of Neonectria are plant pathogens causing cankers and other diseases on hardwood and coniferous trees (Castlebury et al. 2006; Rossman et al. 2008; Crane et al. 2009; Chaverri et al. 2011; Schmitz et al. 2017; Wenneker et al. 2017). Neonectria neomacrospora has been added to the European and Mediterranean Plant Protection Organization (EPPO) alert list (EPPO, 2019).
Classification – Sordariomycetes, Hypocreomycetidae, Hypocreales, Nectriaceae
Type species – Neonectria ramulariae Wollenw., Annls mycol. 15(1/2):52 (1917)
Distribution – Worldwide
Disease symptoms – Canker
Dead shoots can be observed in the lower branches or all over the affected tree. Affected branches or trunks show canker and some may have abundant resin flow. When the canker girdles the affected area, part of the tree above the canker dies. Under humid conditions characteristic small, red fruiting bodies will be formed. Badly affected trees will eventually die (Castlebury et al. 2006).
Beech (Fagus) bark disease is caused by N. coccinea, N. ditissima, N. fuckeliana and N. faginata. Cankers of fruit trees are caused by N. rugulosa and N. ditissima. Shoot dieback of Abies species is caused by N. neomacrospora (Castlebury et al. 2006; Rossman et al. 2008; Crane et al. 2009; Chaverri et al. 2011; Schmitz et al. 2017; Wenneker et al. 2017).
Hosts – Wide host range including plant genera in Amaryllidaceae, Aracaceae, Araliaceae, Betulaceae, Ericaceae, Fagaceae, Lauraceae, Myrtaceae, Pinaceae, Proteaceae, Rosaceae, Sapindaceae and Vitaceae (Farr and Rossman 2019).
Morphological based identification and diversity
The genus Neonectria was established by Wollenweber (1917). The generic concept of Neonectria has been revised by different authors (Booth 1959; Samuels and Brayford 1994; Rossman et al. 1999). Rossman et al. (1999) accepted only three species (N. coccinia, N. galligena, and N. ramulariae) in Neonectria. Subsequently, species were added to the genus based on morphology and/or phylogeny (Hirooka and Kobayashi 2005; Castlebury et al. 2006; Luo and Zhuang 2010a, b; Peng et al. 2011; Lombard et al. 2014, 2015). However, some unrelated species were transferred to other genera based on molecular analyses and morphological data (Lombard et al. 2014, 2015). There are 31 species recognized in the genus, while 23 species have sequence data in GenBank (4/7/2019). Morphological characters (perithecial morphology, ascospore size, macroconidial morphology, presence or absence of microconidia and chlamydospores) along with DNA sequence analysis are appropriate for identification of Neonectria species (Brayford et al. 2004).
Molecular based identification and diversity
Since 2001, DNA sequence analysis has been used to clarify the taxonomy of Neonectria (Mantiri et al. 2001; Brayford et al. 2004; Halleen et al. 2004; Hirooka et al. 2005; Chaverri et al. 2011). Mantiri et al. (2001) and Brayford et al. (2004) used mt SSU rDNA sequence data to infer intrageneric relationships of some Neonectria and Cylindrocarpon species. Later, Halleen et al. (2004) used mt LSU rDNA, TUB2 and nrDNA ITS regions to separate some Cylindrocarpon species included in the N. mammoidea group. Chaverri et al. (2011) approached a comprehensive treatment of Cylindrocarpon and Neonectria based on combined loci analyses and morphological data. Chaverri et al. (2011) defined Neonectria sensu stricto within Nectriaceae with Cylindrocarpon sensu stricto based on multi-gene phylogeny of ITS, LSU, tef1, TUB2, ACT, and RPB1. The ITS, tef1 and TUB2 loci possess highly variable regions (Chaverri et al. 2011) and are important in species delimitation of Neonectria. Rossman et al. (2013) proposed to protect the generic name Neonectria over Cylindrocarpon. Maharachchikumbura et al. (2015) considered Cylindrodendrum not to be congeneric with Neonectria and accepted Neonectria over Cylindrocarpon.
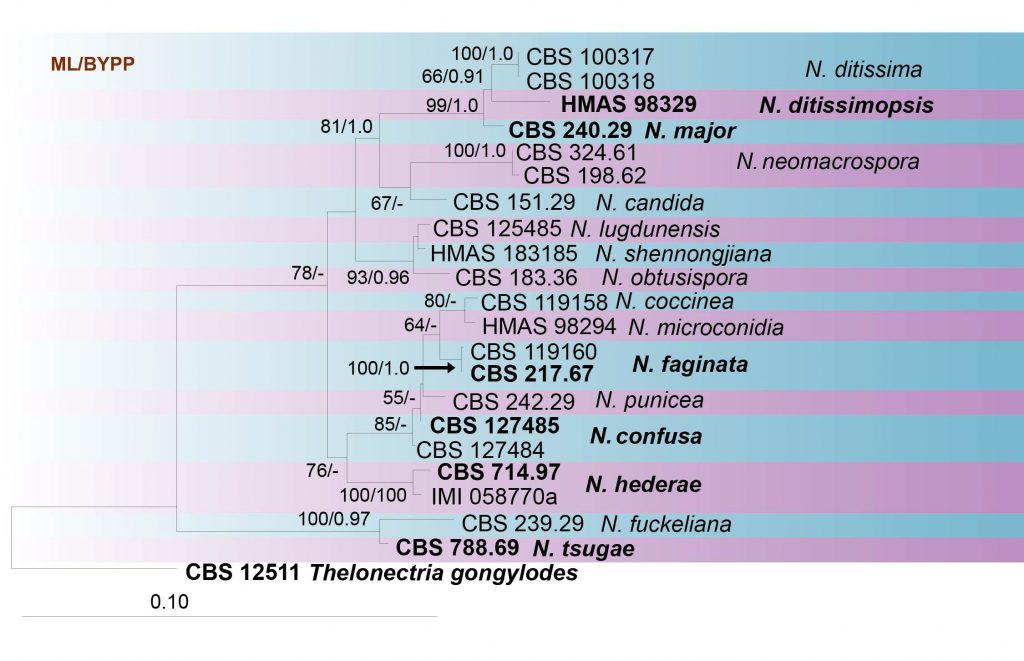
This study reconstructs the phylogeny of Neonectria based on analyses of a combined ITS, LSU, tef1 and TUB2 sequence data (Table 11, Fig. 17). The phylogenetic tree is updated with recently introduced Neonectria species and corresponds to previous studies (Chaverri et al. 2011; Lombard et al. 2014; Mantiri et al. 2001).
Recommended genetic markers (genus level) – LSU, ITS, tef1 and TUB2
Recommended genetic markers (species level) – ITS, tef1 and TUB2
The accepted number of species: 28 species
References: Rossman et al. 1999 (morphology), Brayford et al. 2004; Hirooka and Kobayashi 2007; Chaverri et al. 2011; Lombard et al. 2014 (morphology, phylogeny).
Table. Details of the Neonectria isolates used in the phylogenetic analyses. Ex-type (ex-epitype) strains are in bold and marked with an asterisk* and voucher strains are in bold
| Species | Isolate/Voucher no | ITS | LSU | TUB | tef1 |
| Neonectria austroradicicola | PDD 46334/ G.J.S. 83-154 | EF607077 | – | – | – |
| N. candida | CBS 151.29/ IMI
113894/ MUCL 28083 |
AY677291 | AY677333 | DQ789863 | DQ789723 |
| N. coccinea | CBS 119158/ GJS 98-114 | JF268759 | KC660620 | KC660727 | JF268734 |
| N. confusa | CBS 127485/ HMAS 99197* | FJ560437 | KM515934 | FJ860054 | – |
| N. confusa | CBS 127484/ HMAS 99198 | KM515889 | KM515933 | KM515886 | – |
| N. ditissima | CBS 100318 | KM515890 | KM515935 | DQ789858 | KM515944 |
| N. ditissima | CBS 100317 | KM515891 | KM515936 | KM515887 | KM515945 |
| N. ditissimopsis | HMAS 98329* | JF268764 | – | JF268729 | JF268745 |
| N. faginata | CBS 217.67/ IMI 105738/ ATCC 16547* | HQ840385 | HQ840382 | JF268730 | JF268746 |
| N. faginata | CBS 119160/ GJS 04-159 | HQ840384 | HQ840383 | DQ789883 | DQ789740 |
| N. fuckeliana | CBS 239.29/ IMI 039700 | HQ840386 | HQ840377 | DQ789871 | JF268748 |
| N. hederae | IMI 058770a/ ATCC 16543* | – | KC660617 | DQ789895 | DQ789752 |
| N. hederae | CBS 714.97/ PD 97/1932 | – | KC660616 | DQ789878 | KC660461 |
| N. lugdunensis | CBS 125485/ DAOM 235831/ TG 2008-07 | KM231762 | KM231625 | KM232019 | KM231887 |
| N. major | CBS 240.29/ IMI 113909* | JF735308 | KM515942 | DQ789872 | JF735782 |
| N. microconidia | HMAS 98294 | KC660530 | KC660587 | – | – |
| N. neomacrospora | CBS 198.62/ BBA 9628/ IMI 113890 | AJ009255 | HM364316 | HM352865 | HM364351 |
| N. neomacrospora | CBS 324.61/ DSM 62489/ IMB 9628 | JF735312 | HM364318 | DQ789875 | – |
| N. obtusispora | CBS 183.36/ IMI 113895 | AM419061 | KM515943 | AM419085 | JF735796 |
| N. punicea | CBS 242.29 | KC660522 | KC660565 | DQ789873 | DQ789730 |
| N. shennongjiana | HMAS 183185 | FJ560440 | – | FJ860057 | – |
| N. tsugae | CBS 788.69* | KM231763 | HQ232146 | KM232020 | – |
| Thelonectria gongylodes | CBS 12511/ GJS 90-48 | JQ403330 | JQ403369 | HM352870 | HM364357 |
Fig. Phylogenetic tree generated by maximum likelihood analysis of combined ITS, LSU, tef1 and TUB sequence data of Neonectria species. Related sequences were obtained from GenBank. Twenty-three strains are included in the analyses, which comprise 2336 characters including gaps. Tree topology of the ML analysis was similar to the one generated from BI (figure not shown). The best scoring RAxML tree with a final likelihood value of -7942.756270 is presented. The matrix had 525 distinct alignment patterns, with 18.13% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.223071, C = 0.285136, G = 0.261197, T = 0.230596; substitution rates AC = 1.213729, AG = 2.500008, AT = 1.727890, CG = 0.720430, CT = 6.191594, GT = 1.000000; gamma distribution shape parameter α = 0.749195. Maximum likelihood bootstrap support (≥55%) and posterior probabilities (BYPP≥0.90) from Bayesian inference analysis are indicated respectively near the nodes. Ex-type strains are in bold. The tree is rooted in Thelonectria gongylodes.

No Comments