16 Oct Plasmopara
Plasmopara J. Schröt., in Cohn, Krypt.-Fl. Schlesien 3.1(9–16): 236 (1886) [1889]
The genus Plasmopara belongs to the family Peronosporaceae of the Peronosporales in Oomycetes (Riethmu¨ller et al. 2002; Görg et al. 2017). This genus is included in this study as it is an important plant pathogen on many economically important crops. Plasmopara was introduced by Schröter (1886), but no species was assigned as a type for the genus (Constantinescu et al. 2005). Plasmopara nivea is considered as the type species of this genus (Constantinescu et al. 2005). Plasmopara species are commonly known as downy mildew pathogens. There are 2064 records in USDA fungal database under genus Plasmopara (Farr and Rosman 2019). Downy mildew has become one of the most troublesome diseases in agriculture including P. viticola onthe grape, P. geranii on geranium and P. halstedii on sunflower (McTaggart et al. 2015). Kamoun et al. (2015) categorized P. viticola among the top ten Oomycetes pathogens in plant pathology. The current interest in this genus is to understand the co-evolution with the host and effective disease management (Thines 2010). Taxonomically useful morphological or ecological characters are few for the downy mildews, so as to make identification of synapomorphic states impossible (Göker et al. 2003).
Classification – Oomycota incertae sedis, Peronosporea, Peronosporidae, Peronosporales, Peronosporaceae
Type species – Plasmopara nivea (Unger) J. Schröt.
Distribution – Worldwide
Disease symptoms – Downy mildew
Hosts – Species belonging to this genus are obligate biotrophs on a wide range of hosts including Acanthaceae, Asteraceae, Balsaminaceae, Geraniaceae, Malvaceae, Onagraceae, Orobanchaceae, Violaceae and Vitaceae (Voglmayr et al. 2004; Thines 2010; McTaggart et al. 2015).
Morphological based identification and diversity
Wilson (1907) was the first to consider P. pygmaea as the type of Plasmopara. However, even after a century of discussion of nomenclatural and taxonomic problems of this genus, Plasmopara was contained two different groups by morphology and phylogeny (Constantinescu et al. 2005). Constantinescu et al. (2005) proposed to introduce a new generic name for P. pygmaea and for six other related species and they have established the current classification for Plasmopara.
Species belonging to Plasmopara have the following characters. Hyphae are intercellular, haustoria are intracellular, as obpyriform, globose, or slightly elongated vesicles (Göker et al. 2003; Voglmayr et al. 2004; Constantinescu et al. 2005). A callose sheath often surrounds haustoria. Sporangiophores are mostly present on the underleaf surface of the host, but sometimes also on other parts of the plant (eg. P. viticola produces sporangiophores on inflorescences and youngberries, Zhang et al. 2017). Sporangiophores are colorless, branched in the upper part. They branch monopodially, in two to more orders (Göker et al 2003; Constantinescu et al. 2005). Branches are more or less divergent, ending in a number of elongated ultimate branchlets. The newly formed wall closing the tip after sporangium discharge (Constantinescu et al. 2005). Callose plugs usually present in trunk and/or branches. Sporangiogenesis is holoblastic. These species produce sporangia synchronously, which vary in shape, the sporangia wall is colorless, appearing smooth in light microscopy but showing various types of ornamentations in the electron microscope (Constantinescu et al. 2005). There are 199 epithets listed in Index Fungorum (2019), however, 40 of them do not belong to Plasmopara based on phylogenetic evidence.
Fig. Grape downy mildew disease symptoms. a infected grapevines b-c appearance of oil spot on the upper leaf surface. d sporulation on the lower leaf surface. e young infected berries with sporulation. f. infection on fruits.
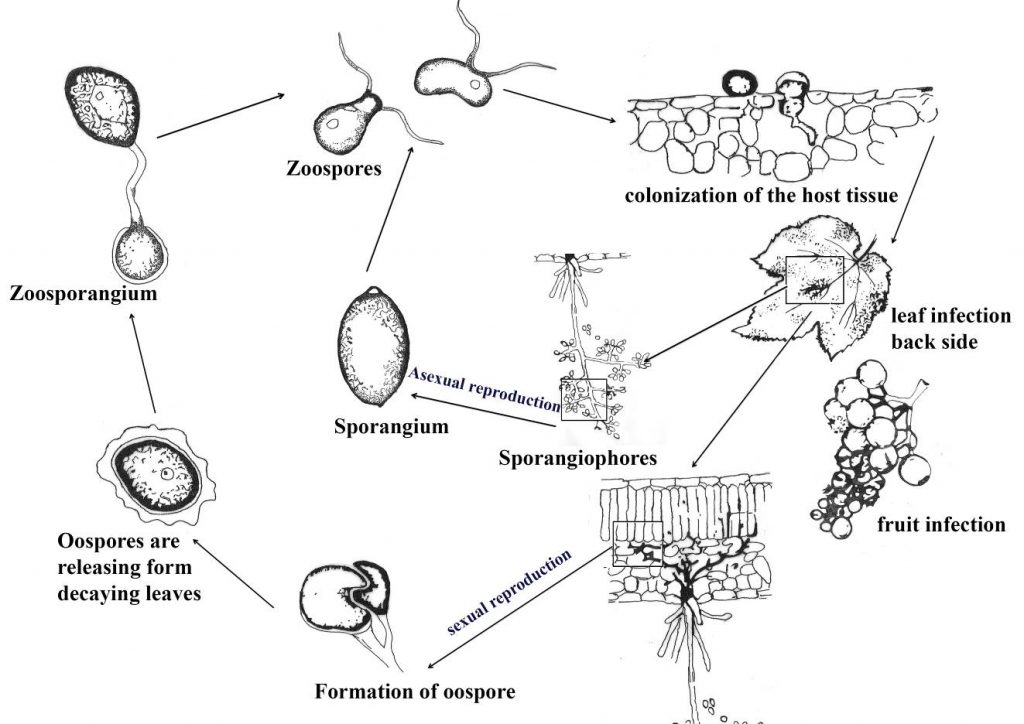
Fig. Disease cycle of grape downy mildew. Redrawn from Kassemeyer et al. (2015)
Oospores develop a single germ tube, terminating with sporangium, once a sporangium disseminated, by rain flash or wind, it releases zoospores. (Ash 2000; Rossi et al. 2007; Carisse 2015; Kamoun et al. 2015; Wilcox 2015).
Molecular based identification and diversity
Molecular and phylogenetic studies have shown that Plasmopara is polyphyletic (Riethmüller et al. 2002; Göker et al. 2003, 2007; Voglmayr et al. 2004; Voglmayr and Contantinescu 2008). Even though Wilson (1907) proposed P. pygmaea as the type species of the genus Plasmopara, it has a close relationship with the Bremia, Paraperonospora, and Basidiophora (Göker et al. 2003). With these morphological and phylogenetic aspects, many species that are traditionally included in Plasmopara have moved into new genera. The newly introduced genera are Viennotia (Göker et al. 2003), Protobremia (Voglmayr et al. 2004), and Plasmoverna (Constantinescu et al. 2005). Constantinescu et al. (2005) resolved Plasmopara phylogeny, introducing Plasmoverna as a new genus to accommodate the morphologically dissimilar and polyphyletic taxa belonging to previous classifications. Voglmayr and Contantinescu (2008) re-classified three species of Plasmopara into new genus Novotelnova Voglmayr & Constant. The species belonging to Novotelnova were identical in the analyses of the nuLSU and nuSSU-ITS1-5.8S datasets. Therefore, in the present study, we follow Voglmayr and Contantinescu (2008) to provide a backbone tree for Plasmopara using combined nuLSU sequence data.
The downy mildew pathogens have been studied extensively to understand their host specificity and co-evolution with the host plants. The grape downy mildew has been identified as a host-specific cryptic species (Rouxel et al 2013; Zhang et al. 2017). Roxuel et al. (2013) considered the cryptic species as formae speciales: P. viticola f. sp. riparia (lineage A occurring on V. riparia and some hybrids); P. viticola f. sp. aestivalis (lineage B found on V. aestivalis, V. labrusca, V. vinifera and some hybrids); P. viticola f. sp. vinifera (lineage C occurring on V. vinifera and some hybrids); P. viticola f. sp. quinquefolia (lineage D found on V. quinquefolia). To understand the cryptic lineages genealogical concordance phylogenetic species recognition (GCPSR) approach is currently accepted (Taylor et al. 2000; Rouxel et al 2013). GCPSR facilitates the most convenient analysis for species that cannot be cultivated or mate in control conditions (O’Donnell et al. 2000; Steenkamp et al. 2002; Roxuel et al 2013).
Recommended genetic markers (genus level) – LSU
Recommended genetic markers (species level) – LSU
The universal barcode for the Oomycetes, the cytochrome oxidase subunit 1 and 2 genes (cox 1 and cox 2) are used. However Choi et al. (2005) suggested cox2 is better suited to this because of its ease of amplification among oomycete lineages, better performance on herbarium specimens, higher discriminatory power at the species level and the availability of a large taxonomically diverse database that already includes many species of oomycetes, especially the downy mildew. In previous studies, nuLSU gene regions (D1–D3 and D7–D8 sequences) were widely used (Riethmüller et al. 2002; Göker et al. 2003; 2007; Voglmayr et al. 2004; Voglmayr and Constantinescu 2008). Branch supports of the backbone tree was often higher in the nuLSU data, which resulted in a larger data matrix and a higher number of parsimony-informative characters than when ITS was used (Voglmayr and Contantinescu, 2008). No study has combined these gene regions or any other gene regions as the marker to understand the phylogenetic relationship within the genus.
The accepted number of species: There are 199 species in Index Fungorum (2019) and only 19 species have molecular data in this genus.
References: Riethmüller et al. 2002; Göker et al. 2003; 2007; Voglmayr et al. 2004; Voglmayr and Constantinescu 2008; Rouxel et al 2013; Zhang et al. 2017 (morphology, phylogeny).
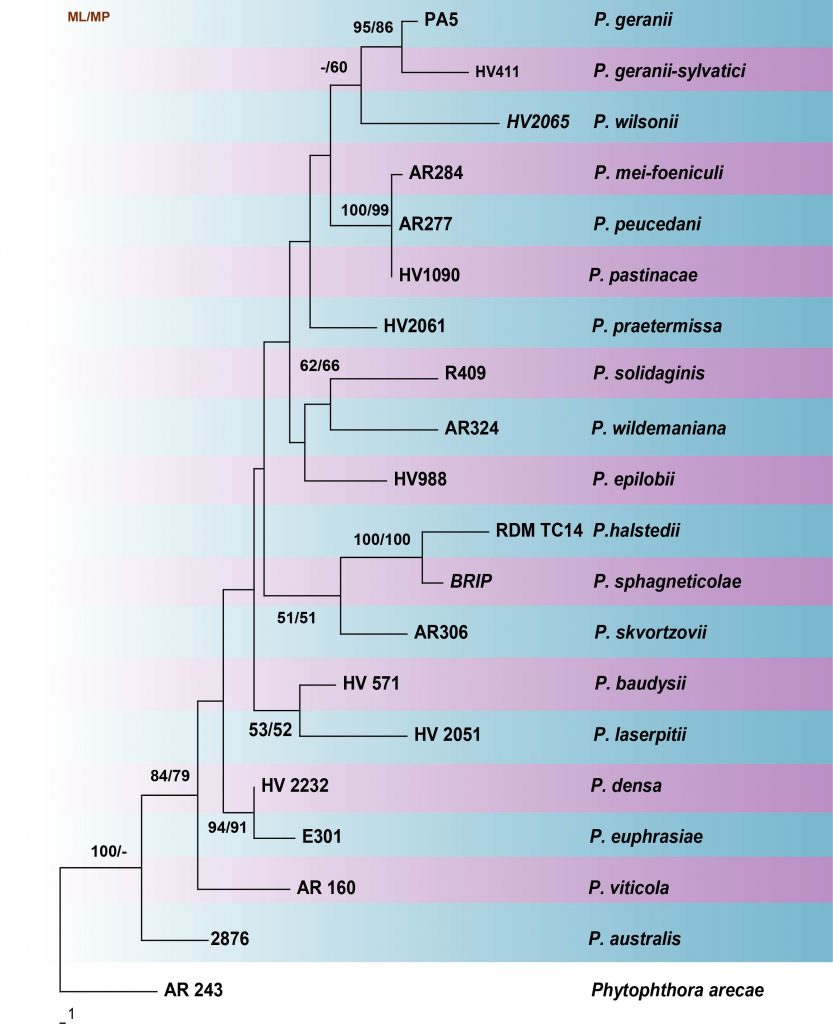
Table Details of the Plasmopara isolates used in the phylogenetic tree
| Species | Isolate/Voucher No | Host | LSU |
| Plasmopara australis | HV 2867 | Luffa cylindrica | KT159461 |
| P. baudysii | HV 571 | Berula erecta | AY035517 |
| P. densa | HV 2232 | Rhinanthus minor | EF553463 |
| P. epilobii | HV988 | Epilobium parviflorum | AY250178 |
| P. euphrasiae | EV 301 | Euphrasia rostkoviana | EF553467 |
| P. geranii | PA5 | Geranium maculatum | DQ148397 |
| P. geranii-sylvatici | HV411 | Geranium sylvaticum | DQ148398 |
| P. halstedii | RDM TC14 | Rudbeckia fulgida | KP164999 |
| P. laserpitii | HV2051 | Laserpitium latifolium | KC495034 |
| P. mei-foeniculi | AR284 | Meum athamanticum | AY250160 |
| P. pastinacae | HV1090 | Pastinaca sativa | AY250157 |
| P. peucedani | AR277 | Peucedanum palustre | AY250154 |
| P. praetermissa | HV2061 | Geranium sylvaticum | DQ148396 |
| P. skvortzovii | AR306 | Abutilon theophrasti | AY250179 |
| P. solidaginis | R409 | Solidago virgaurea | AY250144 |
| P. sphagneticolae | BRIP | Sphagneticola trilobata | KM085176 |
| P. viticola | AR 160 | Vitis vinifera | AY035524 |
| P. wildemaniana | AR324 | Hypoestes sp. | AY250180 |
| P. wilsonii | HV2065 | Geranium nepalense | DQ148408 |
| Phytophthora arecae | AR 243 | Unknown | AY035530 |
Fig. Phylogenetic tree generated by maximum parsimony analysis of LSU sequence data of Plasmopara species. Related sequences were obtained from GenBank. Nineteen strains are included in the analyses, which comprise 1263 characters including gaps. The tree was rooted with Phytophthora arece (AR 234). Tree topology of the MP analysis was similar to the ML. The maximum parsimonious dataset consisted of constant 1053, 110 parsimony-informative and 100 parsimony-uninformative characters. The parsimony analysis of the data matrix resulted in the maximum of two equally most parsimonious trees with a length of 365 steps (CI = 0.641, RI = 0.539, RC = 0.345, HI = 0.359) in the first tree. The best scoring RAxML tree with a final likelihood value of -3660.849005 is presented. The matrix had 254distinct alignment patterns, with 30.91% of undetermined characters or gaps. Estimated base frequencies were as follows; A = 0.229156, C = 0.177763, G = 0.308486, T = 0.284595; substitution rates AC = 0.706031, AG = 4.368078, AT = 1.048734, CG = 0.141039, CT = 7.063364, GT = 1.000000; gamma distribution shape parameter α = 0.163551. RAxML and maximum parsimony bootstrap support value ≥ 50% (BT) are shown respectively near the nodes.


No Comments