01 Nov Pyrenophora
Pyrenophora
Background
Pyrenophora represents a genus of plant pathogenic fungi associated with a wide variety of substrates. Fries (1849) list the genus as Pyrenophora typified with Pyrenophora phaeocomes. The genus Pyrenophora clusters in the suborder Pleosporineae of the family Pleosporaceae (Berbee 1996; Zhang and Berbee 2001; Hyde et al. 2013a, b; Zhang et al. 2012; Ariyawansa et al. 2014). Recent studies using multi-gene analysis and some coupled with morphology have provided the groundwork for classification of species in Pyrenophora (Berbee 1996; Zhang and Berbee 2001; Hyde et al. 2013a, b; Zhang et al. 2012).
Pyrenophora has been linked to asexual morphs in Drechslera. Pyrenophora species are important plant pathogens as well as saprobes. Many species cause disease on their graminivorous hosts and are usually present in their asexual state (Drechslera) (Zhang and Berbee 2001). Species of Pyrenophora are serious plant pathogens (Zhang and Berbee 2001). Pyrenophora teres (Drechslera teres) is a necrotrophic pathogen of economically important crops, such as barley (Gupta and Loughman 2001; Kingsland 1991). Pyrenophora graminea (Drechslera graminea) causes barley stripe resulting in significant yield losses (Tekauz 1983, 1990). Pyrenophora graminea lives within barley kernels as mycelium, and when seeds germinate, hyphae enter the seedling through the coleorhiza, causing a systemic infection (Platenkamp 1976; Porta-Puglia et al. 1986). Pyrenophora tritici-repentis causes a tan spot of wheat (Lamari and Bernier 1989) which occurs in all the major wheat-growing areas of the world and causes 3 to 50 % yield losses (Ballance et al. 1996). Its prevalence has increased recently.
Some Pyrenophora species have been used as biocontrol agents. Bromus tectorum is a dominant winter annual weed in cold deserts of the western United States (Meyer et al. 2007). Together with other annual brome grasses, it has invaded many ecosystems of the western United States creating near-monocultures in which the native vegetation cannot compete (Meyer et al. 2007). Pyrenophora semeniperda has to be used as a biocontrol agent to kill the dormant seeds of Bromus tectorum (Meyer et al. 2007). Several studies have assessed chemical production by Pyrenophora species. A new phytotoxic sesquiterpenoid Penta-2,4-dienoic acid (pyrophoric acid) was isolated from solid wheat seed culture of P. semeniperda.
Species identification and numbers
Pyrenophora is characterized by immersed, erumpent to nearly superficial ascomata, indefinite pseudoparaphyses, clavate to saccate asci usually with a large apical ring, and muriform terete ascospores. Morphologically, the terete ascospores of Pyrenophora can be easily distinguished from Clathrospora and Platyspora. The indefinite pseudoparaphyses and smaller ascospores of Pyrenophora can be clearly separated from those of Pleospora (Sivanesan 1984). Pyrenophora species can easily be distinguished from species in Cochliobolus and Setosphaeria on the basis of the shape, septation, and color of the ascospores (Zhang and Berbee 2001). Drechslera species were initially categorized in Helminthosporium on the basis of their dark color, transversely septate conidia and a graminivorous habitat (Shoemaker 1959). Consequently, graminivorous Helminthosporium species were segregated into three genera, Bipolaris, Drechslera, and Exserohilum, defined based on their association with their sexual states Cochliobolus, Pyrenophora, or Setosphaeria, respectively (Zhang and Berbee 2001). Currently, 198 species of Pyrenophora and 135 species of Drechslera are listed in Index Fungorum (2014).
Molecular phylogeny
Rapid identification of diseases caused by Pyrenophora has been determined via different DNA markers. Identification of molecular genetic markers in Pyrenophora teres f. teres associated with low virulence on ‘Harbin’ barley was assessed by random amplified polymorphic DNA (RAPD) (Weiland et al. 1999) and five RAPD markers were obtained that were associated in coupling with low virulence. The data suggested that the RAPD technique can be used to tag genetic determinants for virulence in P. teres f. teres (Weiland et al. 1999). Specific polymerase chain reaction (PCR) primers were developed from amplified fragment length polymorphism (AFLP) fragments of P. teres, in order to distinguish the two forms, P. teres f. teres (which cause net form blotch on barley leaves) and P. teres f. maculata (which causes spot form); the two forms are morphologically very similar in culture (Leisova et al. 2005). The PCR assay was certified with 60 samples of Pyrenophora species. The amplification with four designed PCR primer pairs provided P. teres form-specific products. No cross-reaction was observed with the DNA of several other species, such as P. tritici-repentis and P. graminea (Leisova et al. 2005). Pyrenophora graminea is the causal agent of barley leaf stripe disease (Mokrani et al. 2012). Two leaf stripe isolates PgSy3 (exhibiting high virulence on the barley cultivar ‘Arabi Abiad’) and PgSy1 (exhibiting low virulence on Arabi Abiad), were mated and 63 progeny were isolated and phenotyped for the reaction on Arabi Abiad (Mokrani et al. 2012). From 96 AFLP markers, three AFLP markers, E37M50-400, E35M59-100, and E38M47-800 were linked to the virulence locus VHv1 in isolate PgSy3. Lubna et al. (2012) suggested that the three markers are closely linked to VHv1 and are unique to isolates carrying the virulence locus. Pecchia et al. (1998) developed an efficient PCR protocol for amplification of the IGS region in P. graminea and to characterize this region by restriction fragment analysis. During the study based on the length of the IGS-PCR product, ca. 3.8 or 4.4 kb, two groups of isolates were identified from six cultures i.e. I3/88 (Italy; CBS 100862), I7/88 (Italy; CBS100861), 60/ 93 (Austria; CBS 100866), I10/95 (Tunisia; CBS 100863), I28/95 (Tunisia; CBS 100864), I33/95 (Tunisia; CBS 100865). The RFLP patterns of isolates obtained with the 6- base cutting enzymes ApaI, BglII, DraI, EcoRV, HindIII and SacI were similar within each group and different between the two groups (Pecchia et al. 1998). Restriction patterns of IGS- PCR products digested with the 4-base cutting enzyme AluI were polymorphic among isolates in spite of their IGS-PCR product length (Pecchia et al. 1998).
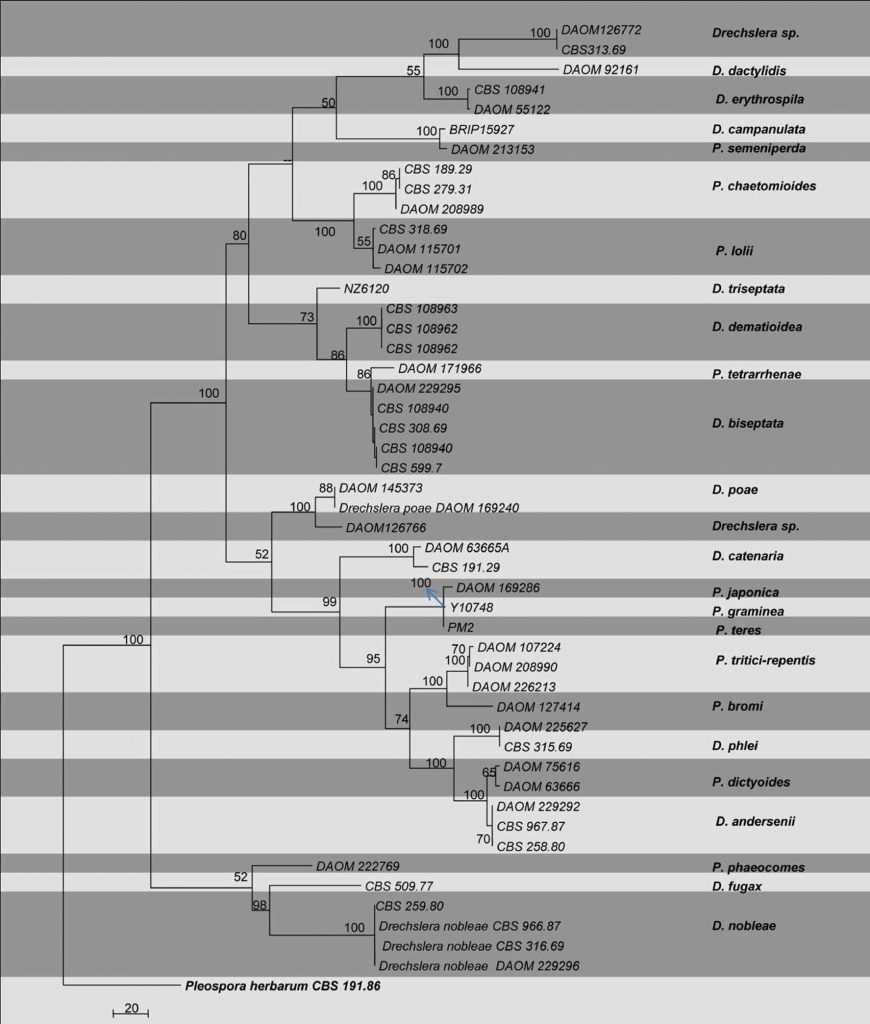
Molecular studies of Pyrenophora/Drechslera species have detailed the taxonomic placement of the genus. Initially, the 18S rRNA gene was used for the classification of Pyrenophora/Drechslera and related genera (Berbee 1996). Phylogenetic analysis based on 18S rRNA showed Pyrenophora to cluster within the Pleosporaceae (Zhang and Berbee 2001) rather than in Pyrenophoraceae (Zhang and Berbee 2001). Later, phylogenetic analysis of the ITS and gdp data showed that Pyrenophora is monophyletic (Zhang and Berbee 2001), and the asexual state Drechslera clustered with their predicted sexual relatives (Table, Fig.).
Table Pyrenophora. Details of the isolates used in the phylogenetic tree
| Species | Isolate | GenBank association numbers | |||
| ITS | LSU | GPDH | |||
| Drechslera andersenii | CBS 258.80 | AY004804 | AY004835 | ||
| D. andersenii | CBS 967.87 | AY004805 | |||
| D. andersenii | DAOM 229292 | JN943646 | JN940084 | ||
| D. avenae | CBS 189.29 | AY004795 | AY004827 | ||
| D. avenae | CBS 279.31 | AY004796 | AY004828 | ||
| D. biseptata | DAOM 208987 | AY004786 | AY004817 | ||
| D. biseptata | CBS 308.69 | JN712464 | JN712530 | AY004819 | |
| D. biseptata | CBS 599.7 | AY004787 | AY004818 | ||
| D. biseptata | CBS 108940 | AY004788 | |||
| D. campanulata | BRIP15927 | AF163058 | |||
| D. catenaria | DAOM 63665A | AY004802 | AY004833 | ||
| D. catenaria | CBS 191.29 | AY004803 | AY004834 | ||
| D. dactylidis | DAOM 92161 | AY004781 | AY004812 | ||
| D. dematioidea | CBS 108963 | AY004789 | JN712532 | AY004820 | |
| D. dematioidea | DAOM 229295 | JN943648 | JN940094 | ||
| D. dematioidea | CBS 108962 | JN712465 | JN712531 | ||
| D.dematioidea | CBS 108962 | AY004790 | JN712531 | AY004821 | |
| Drechslera dictyoides | DAOM 63666 | AY004806 | JN940080 | AY004836 | |
| D. erythrospila | CBS 108941 | AY004782 | AY004813 | ||
| D. erythrospila | DAOM 55122 | AY004783 | AY004814 | ||
| D. fugax | CBS 509.77 | AY004791 | AY004822 | ||
| D.nobleae | CBS 259.80 | AY004792 | AY004823 | ||
| D. nobleae | DAOM 229296 | JN943647 | JN940095 | ||
| D. nobleae | CBS 966.87 | AY004793 | AY004824 | ||
| D. nobleae | CBS 316.69 | AY004794 | AY004825 | ||
| D. phlei | CBS 315.69 | AY004807 | AY004837 | ||
| D. phlei | DAOM 225627 | JN943656 | JN940077 | ||
| D. poae | DAOM 145373 | AY004801 | JN940082 | AY004832 | |
| D. poae | DAOM 169240 | JN943651 | |||
| D. siccans | DAOM 115701 | AY004797 | JN940078 | ||
Ex-type (ex-epitype) strains are bolded and marked with an * and voucher stains are bold.
Fig. Phylogram generated from parsimony analysis based on combined ITS, gdp and LSU sequenced data of Pyrenophora. Parsimony bootstrap support values greater than 50 % are indicated above the nodes. The ex-type (ex-epitype) and voucher strains are in bold. The tree is rooted with Pleospora herbarium CBS 276.37
Recommended genetic markers
- Large small subunits of nrDNA (LSU)–generic level
- ITS and gdp–inter-specific delineation
Based on our phylogeny, we observed that gdp gives high resolution compared to ITS and LSU, such that it can be readily used to determine the placement of Pyrenophora species.

No Comments